For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ITATDLEVRAGARTLVYAPGPALRIQPGDRIGLVGRNGAGKTTSMRILAGEGEPYAGSVSRIGEIGYLPQ DPKEGNLDLLARDRVLSARGLDTIISEMEKQQTLMAELADDDQRDKAVRRYGQLEERFSSLGGYVAESEA ARICSSLGLPERILVQPLRTLSGGQRRRVELARILFGASEGGAGSSMTLLLDEPTNHLDADSIGWLRSFL QNHDGGLVVISHNVELLADVVNKVWFLDAVRGEVDVYNMGWQKYLDARSLDEQRRRRERANAEKKASALR TQAAKMGAKATKAVAAQNMLRRAEKMLSGLDEERVADKVARIKFPTPAACGRVPLVAKGLTKNYGSLEIF TGVDLAIDRGSRVVVLGLNGAGKTTLLRLLAGVETADAGELEPGHGLKLGYFAQEHDTLDDHATVWENIR HAAPDTGEQELRGILGAFMFSGPQLEQLAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQV LDALRSYLGAVVLVTHDPGAAEALNPQRVVLLPDGTEDYWSDEYRDLIELA*
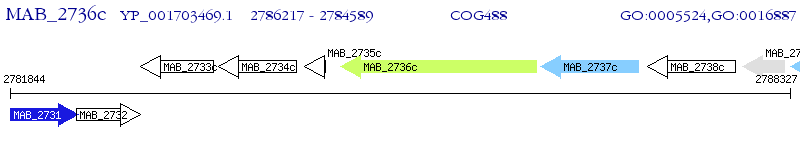
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2736c | - | - | 100% (542) | macrolide ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1560 | - | 5e-61 | 31.05% (512) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1846 | - | 3e-55 | 32.74% (559) | putative ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1508 | - | 0.0 | 84.32% (542) | macrolide ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_3668 | - | 0.0 | 87.45% (542) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1473 | - | 0.0 | 84.32% (542) | macrolide ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_01816 | - | 0.0 | 82.20% (545) | ABC transporter ATP-binding protein, possibly in EF-3 subfamily |
| M. marinum M | MMAR_2279 | - | 0.0 | 85.42% (542) | ATP-binding protein ABC transporter |
| M. avium 104 | MAV_3306 | - | 0.0 | 84.77% (545) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_3140 | - | 0.0 | 87.64% (542) | ABC transporter ATP-binding protein |
| M. thermoresistible (build 8) | TH_3270 | - | 0.0 | 85.14% (545) | PUTATIVE ABC transporter-related protein |
| M. ulcerans Agy99 | MUL_1481 | - | 0.0 | 85.24% (542) | ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2742 | - | 0.0 | 87.08% (542) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2279|M.marinum_M ---------------------MITATDLEVRAGARILLSP-DGPDLRVQP
MUL_1481|M.ulcerans_Agy99 ---------------------MITATDVEVRAGARILLSP-DGPDLRVQP
MAV_3306|M.avium_104 ---------------------MITATDLEVRAGARILLSP-DGPDLRVQP
MLBr_01816|M.leprae_Br4923 ---------------------MITATDLEVRAGARILLSS-DGPDLRVQP
Mb1508|M.bovis_AF2122/97 ---------------------MITATDLEVRAGARILLAP-DGPDLRVQP
Rv1473|M.tuberculosis_H37Rv ---------------------MITATDLEVRAGARILLAP-DGPDLRVQP
Mflv_3668|M.gilvum_PYR-GCK ---------------------MITATDLEVRAGARTLLSV-EGAALRVQP
Mvan_2742|M.vanbaalenii_PYR-1 MPTPAWVPVQPCHAYLTRSISVITATDLEVRAGARTLLSI-EGSALRVQP
MSMEG_3140|M.smegmatis_MC2_155 ---------------------MITATDLEVRAGARTLLLI-EGSALRVQP
TH_3270|M.thermoresistible__bu ---------------------VITASGLEVRAGARTLLSLDDGQVLRVQP
MAB_2736c|M.abscessus_ATCC_199 ---------------------MITATDLEVRAGARTLVYA-PGPALRIQP
:***:.:******* *: * **:**
MMAR_2279|M.marinum_M GDRIGLVGRNGAGKTTTLRILAGESEPYAGSVTRAGEIGYLPQDPKEGDL
MUL_1481|M.ulcerans_Agy99 GDRIGLVGRNGAGKTTTLRILAGESEPYAGSVTRAGEIGYLPQDPKEGDL
MAV_3306|M.avium_104 GDRIGLVGRNGAGKTTTLRILAGETEPYAGTIARSGEIGYLPQDPKEGDL
MLBr_01816|M.leprae_Br4923 GDRIGLVGRNGAGKTTTLRILAGATEPYAGSVTWTGEIGYLPQDPKVGNL
Mb1508|M.bovis_AF2122/97 GDRIGLVGRNGAGKTTTLRILAGEVEPYAGSVTRAGEIGYLPQDPKVGDL
Rv1473|M.tuberculosis_H37Rv GDRIGLVGRNGAGKTTTLRILAGEVEPYAGSVTRAGEIGYLPQDPKVGDL
Mflv_3668|M.gilvum_PYR-GCK GDRIGLVGRNGAGKTTTMRILAGEGEPYAGSVTRTGEIGYLPQDPKEGDL
Mvan_2742|M.vanbaalenii_PYR-1 GDRIGLVGRNGAGKTTTMRILAGEGEPYAGSVTRTGEIGYLPQDPKEGDL
MSMEG_3140|M.smegmatis_MC2_155 GDRIGLVGRNGAGKTTTMRILAGEGEPYAGSVTRSGDIGYLPQDPKEGDL
TH_3270|M.thermoresistible__bu GDRIGLVGRNGAGKTTTLRILAGEGEPYAGTVTRTGDIGYLPQDPKQGDL
MAB_2736c|M.abscessus_ATCC_199 GDRIGLVGRNGAGKTTSMRILAGEGEPYAGSVSRIGEIGYLPQDPKEGNL
****************::***** *****::: *:********* *:*
MMAR_2279|M.marinum_M EVLARDRVLSARGLDVLLTDLEKQQALMAEVADDAARDRAIRRYGQLEER
MUL_1481|M.ulcerans_Agy99 EVLARDRVLSARGLDVLLTDLEKQQALMAEVADDAARDRAIRRYGQLEER
MAV_3306|M.avium_104 DVLARDRVLSARGLDVLLTDLEKQQALMAEVADDDARDRAIRRYGQLEER
MLBr_01816|M.leprae_Br4923 DVLARDRVLSARGLDVLLTDLEKQQALMAEVADDDARDRAIRRYGQLEER
Mb1508|M.bovis_AF2122/97 DVLARDRVLSARGLDVLLTDLEKQQALMAEVADEDERDRAIRRYGQLEER
Rv1473|M.tuberculosis_H37Rv DVLARDRVLSARGLDVLLTDLEKQQALMAEVADEDERDRAIRRYGQLEER
Mflv_3668|M.gilvum_PYR-GCK DMLARDRVLSARGLDTLLSDLEKQQALMAEVADDAARDKAVRRYGQLEER
Mvan_2742|M.vanbaalenii_PYR-1 DMLARDRVLSARGLDTLLSDLEKQQALMAEVADDAARDKAVRRYGQLEER
MSMEG_3140|M.smegmatis_MC2_155 DVLARDRVLSARGLDTLLADLEKQQALMAEVADDAARDKAVRRYGQLEER
TH_3270|M.thermoresistible__bu DVLARDRVLSARGLDTLLAELEKQQVLMAEVVDDAARDRAVRRYGQLEER
MAB_2736c|M.abscessus_ATCC_199 DLLARDRVLSARGLDTIISEMEKQQTLMAELADDDQRDKAVRRYGQLEER
::*************.:::::****.****:.*: **:*:*********
MMAR_2279|M.marinum_M FVALGGYGAESEASRICASLGLPERVLTQQLRTLSGGQRRRVELARILFA
MUL_1481|M.ulcerans_Agy99 FVTLGGYGAESEASRICASLGLPERVLTQQLRTLSGGQRRRVELARILFA
MAV_3306|M.avium_104 FVALGGYGAESEASRICASLGLPERVLTQQLRTLSGGQRRRVELARILFA
MLBr_01816|M.leprae_Br4923 FLALGGYSAESEAGRICASLGLPERVLVQQLCTLSGGQRRRVELARILFA
Mb1508|M.bovis_AF2122/97 FVALGGYGAESEAGRICASLGLPERVLTQRLRTLSGGQRRRVELARILFA
Rv1473|M.tuberculosis_H37Rv FVALGGYGAESEAGRICASLGLPERVLTQRLRTLSGGQRRRVELARILFA
Mflv_3668|M.gilvum_PYR-GCK FAALGGYAAESEAGRICASLGLPDRVLTQPLRTLSGGQRRRVELARILFA
Mvan_2742|M.vanbaalenii_PYR-1 FAALGGYAAESEAGRICASLGLPDRVLTQPLRTLSGGQRRRVELARILFA
MSMEG_3140|M.smegmatis_MC2_155 FSALGGYAAESEAGRICASLGLPERVLTQPLHTLSGGQRRRVELARILFA
TH_3270|M.thermoresistible__bu FAALGGYAAESEAGRICASLGLPDRVLTQPLRTLSGGQRRRVELARILFA
MAB_2736c|M.abscessus_ATCC_199 FSSLGGYVAESEAARICSSLGLPERILVQPLRTLSGGQRRRVELARILFG
* :**** *****.***:*****:*:*.* * *****************.
MMAR_2279|M.marinum_M ASD---TGAGSSTVLLLDEPTNHLDADSVGWLRDFLRAHTGGLIIISHNV
MUL_1481|M.ulcerans_Agy99 ASD---TGAGSSTVLLLDEPTNHLDADSVGWLRDFLRAHTGGLIIISHNV
MAV_3306|M.avium_104 AAEGGATGSGSATTLLLDEPTNHLDADSIGWLRDFLRAHTGGLVIISHNV
MLBr_01816|M.leprae_Br4923 ASRAGTGASGSGTTLLLDEPTNHLDADSLGWLRDFLRSHTGGLVVISHNV
Mb1508|M.bovis_AF2122/97 ASE---SGAGNSTTLLLDEPTNHLDADSLGWLRDFLRLHTGGLVVISHNV
Rv1473|M.tuberculosis_H37Rv ASE---SGAGNSTTLLLDEPTNHLDADSLGWLRDFLRLHTGGLVVISHNV
Mflv_3668|M.gilvum_PYR-GCK ASEG-GGG--DATTLLLDEPTNHLDADSIGWLRAFLQNHNGGLVVISHDV
Mvan_2742|M.vanbaalenii_PYR-1 ASEA-GSG--SATTLLLDEPTNHLDADSIGWLRTFLQNHTGGLVVISHDV
MSMEG_3140|M.smegmatis_MC2_155 ASET-GSG--SATTLLLDEPTNHLDADSIGWLRDFLKNHTGGLVMISHNV
TH_3270|M.thermoresistible__bu AADS-GSGGGAGTTLLLDEPTNHLDADSIGWLRTFLQNHTGGLVIISHNV
MAB_2736c|M.abscessus_ATCC_199 ASE---GGAGSSMTLLLDEPTNHLDADSIGWLRSFLQNHDGGLVVISHNV
*: . . .**************:**** **: * ***::***:*
MMAR_2279|M.marinum_M ELLADVVNRVWFLDAVRGEVDVYNMGWHKYLDARATDEQRRRRERANAER
MUL_1481|M.ulcerans_Agy99 ELLADVVNRVWFLDAVRGEVDVYNMGWHKYLDARATDEQRRRRERANAER
MAV_3306|M.avium_104 ELLADVVNRVWFLDAVRGEVDVYNMSWQKYLDARATDEQRRRRERANAER
MLBr_01816|M.leprae_Br4923 ELIAAVVNRVWFLDAVLGKVDVYNMGWYKYLDSRATDEQRRRRERVNAER
Mb1508|M.bovis_AF2122/97 DLVADVVNKVWFLDAVRGQVDVYNMGWQRYVDARATDEQRRIRERANAER
Rv1473|M.tuberculosis_H37Rv DLVADVVNKVWFLDAVRGQVDVYNMGWQRYVDARATDEQRRIRERANAER
Mflv_3668|M.gilvum_PYR-GCK ELLADVVNRVWFLDAVRGEADVYNMGWQKYLDARATDEQRRRRERANAEK
Mvan_2742|M.vanbaalenii_PYR-1 DLLADVVNRVWFLDAVRGEADVYNMGWQKYLDARATDEQRRRRERANAEK
MSMEG_3140|M.smegmatis_MC2_155 ELLEEVVNRVWFLDAVRGEVDVYNMGWQKYLDARATDEQRRRRERANAEK
TH_3270|M.thermoresistible__bu ELLAEVVNRVWFLDAVRGEVDIYNMGWHKYLDARATDEQRRRRERANAER
MAB_2736c|M.abscessus_ATCC_199 ELLADVVNKVWFLDAVRGEVDVYNMGWQKYLDARSLDEQRRRRERANAEK
:*: ***:******* *:.*:***.* :*:*:*: ***** ***.***:
MMAR_2279|M.marinum_M KATALRTQAAKMGAKATKAVAAQNMLRRADRMMAALDEERVADKVARIKF
MUL_1481|M.ulcerans_Agy99 KATALRTQAAKMGAKATKAVAAQNMLRRADRMMAALDEERVADKVARIKF
MAV_3306|M.avium_104 KAAALRTQAAKLGAKATKAVAAQNMLRRADRMMAALDEERVADKVARIKF
MLBr_01816|M.leprae_Br4923 KATALRTQAAKMGAKATKAVAAQNMLRRADRMMAALDEERVIEKVAQIKF
Mb1508|M.bovis_AF2122/97 KAAALRAQAAKLGAKATKAVAAQNMLRRADRMMAALDEERVADKVARIKF
Rv1473|M.tuberculosis_H37Rv KAAALRAQAAKLGAKATKAVAAQNMLRRADRMMAALDEERVADKVARIKF
Mflv_3668|M.gilvum_PYR-GCK KAGALRAQAAKMGAKATKAVAAQNMLRRAERMMAELDAERVADKVARIKF
Mvan_2742|M.vanbaalenii_PYR-1 KASALRAQAAKMGAKATKAVAAQNMLKRAERMMAALDDERVADKVARIKF
MSMEG_3140|M.smegmatis_MC2_155 KAGALRAQAAKMGAKATKAVAAQNMLRRAERMMAALDEERVADKVARIKF
TH_3270|M.thermoresistible__bu KAAALRAQAAKMGAKATKAVAAQNMLRRAERMLAALDDAPVADKVARIKF
MAB_2736c|M.abscessus_ATCC_199 KASALRTQAAKMGAKATKAVAAQNMLRRAEKMLSGLDEERVADKVARIKF
** ***:****:**************:**::*:: ** * :***:***
MMAR_2279|M.marinum_M PTPAACGRVPLVVKGLSKTYGSLEVFTGVDLAIDRGSRVVVLGLNGAGKT
MUL_1481|M.ulcerans_Agy99 PTPAACGRVPLVVKGLSKTYGSLEVFTGVDLAIDRGSRVVVLGLNGAGKT
MAV_3306|M.avium_104 PTPAACGRTPLTATGLSKSYGSLEVFSGVDLAIDRGSRVVVLGLNGAGKT
MLBr_01816|M.leprae_Br4923 PTPAACGRTPLVAKGLSKTYGSLEVFTGVNLAIDRGSRVVVLGLNGAGKT
Mb1508|M.bovis_AF2122/97 PTPAACGRTPLVANGLGKTYGSLEVFTGVDLAIDRGSRVVILGLNGAGKT
Rv1473|M.tuberculosis_H37Rv PTPAACGRTPLVANGLGKTYGSLEVFTGVDLAIDRGSRVVILGLNGAGKT
Mflv_3668|M.gilvum_PYR-GCK PTPAPCGKTPLIAKGLTKTYGSLEIFTGLDLAIDRGSRVVVLGLNGAGKT
Mvan_2742|M.vanbaalenii_PYR-1 PTPAPCGKTPLTAKGLTKTYGSLEIFSGLDLAIDRGSRVVVLGLNGAGKT
MSMEG_3140|M.smegmatis_MC2_155 PTPAPCGRTPLVAKGLTKNYGSLEVFTGVDLAIDRGSRVVVLGLNGAGKT
TH_3270|M.thermoresistible__bu PTPAPCGRTPLVAKGLTKTYGSLEVFTGVDLAIDRGSRVVVLGLNGAGKT
MAB_2736c|M.abscessus_ATCC_199 PTPAACGRVPLVAKGLTKNYGSLEIFTGVDLAIDRGSRVVVLGLNGAGKT
****.**:.** ..** *.*****:*:*::**********:*********
MMAR_2279|M.marinum_M TLLRLLAGVEHPDTGGLEPGHGLRIGYFAQEHDTLDNDATVWENIRHAAP
MUL_1481|M.ulcerans_Agy99 TLLRLLAGVEHPDTGGLEPGHGLRIGYFAQEHDTLDNDATVWENIRHAAP
MAV_3306|M.avium_104 TLLRLLAGAETPDTGRIEPGHGLRIGYFAQEHDTLDNDATVWQNIRHAAP
MLBr_01816|M.leprae_Br4923 TLLRVLAGAEKPDVGRLEPGHGLRIGYFAQEHDTLDNEDTVWQNIRHTAP
Mb1508|M.bovis_AF2122/97 TLLRLLAGVEQPDTGVLEPGYGLRIGYFAQEHDTLDNDATVWENVRHAAP
Rv1473|M.tuberculosis_H37Rv TLLRLLAGVEQPDTGVLEPGYGLRIGYFAQEHDTLDNDATVWENVRHAAP
Mflv_3668|M.gilvum_PYR-GCK TLLRLLAGVEIADAGGLEPGHGLKIGYFAQEHDTLDTMRTVWENIRSAAP
Mvan_2742|M.vanbaalenii_PYR-1 TLLRLLAGVEKADAGEIEPGHGLKIGYFAQEHDTLDNMASVWENIRHAAP
MSMEG_3140|M.smegmatis_MC2_155 TLLRLIAGTEKPDAGHLEPGHGLKIGYFAQEHDTLDNQATVWENIRHAAP
TH_3270|M.thermoresistible__bu TLLRLLAGVETPDAGQIEPGHGLKIGYFAQEHDTLDDHATVWENIRHAAP
MAB_2736c|M.abscessus_ATCC_199 TLLRLLAGVETADAGELEPGHGLKLGYFAQEHDTLDDHATVWENIRHAAP
****::**.* .*.* :***:**::*********** :**:*:* :**
MMAR_2279|M.marinum_M ESGEQELRGLLGAFMFTGPQLDQPAGTLSGGEKTRLALAGLVASTANVLL
MUL_1481|M.ulcerans_Agy99 ESGEQELRGLLGAFMFTGPQLDQPAGTLSGGEKTRLALAGLVASTANVLL
MAV_3306|M.avium_104 DSGEQDLRGLLGAFMFSGPQLDQPAGTLSGGEKTRLALAGLVASTANVLL
MLBr_01816|M.leprae_Br4923 EFGEQDLRGLLGAFMFSGPQLDQLAGTLSGGEKTRLALAGLVASTANVLL
Mb1508|M.bovis_AF2122/97 DAGEQDLRGLLGAFMFTGPQLEQPAGTLSGGEKTRLALAGLVASTANVLL
Rv1473|M.tuberculosis_H37Rv DAGEQDLRGLLGAFMFTGPQLEQPAGTLSGGEKTRLALAGLVASTANVLL
Mflv_3668|M.gilvum_PYR-GCK DTGEQDLRGLLGAFMFTGPQLEQPAGTLSGGEKTRLALAGLVASTANVLL
Mvan_2742|M.vanbaalenii_PYR-1 DTGEQDLRGLLGAFMFTGPQLDQPAGTLSGGEKTRLALAGLVASTANVLL
MSMEG_3140|M.smegmatis_MC2_155 DTGEQDLRGLLGAFMFTGPQLDQPAGTLSGGEKTRLALAGLVASTANVLL
TH_3270|M.thermoresistible__bu DSGEQDLRGLLGAFMFSGPQLEQPAGTLSGGEKTRLALAGLVASAANVLL
MAB_2736c|M.abscessus_ATCC_199 DTGEQELRGILGAFMFSGPQLEQLAGTLSGGEKTRLALAGLVASTANVLL
: ***:***:******:****:* ********************:*****
MMAR_2279|M.marinum_M LDEPTNNLDPASREQVLDALRSYQGAVVLVTHDPGAAEALDPQRVVLLPD
MUL_1481|M.ulcerans_Agy99 LDEPTNNLDPASREQVLDALRSYQGAVVLVTHDPGAAEALDPQRVVLLPD
MAV_3306|M.avium_104 LDEPTNNLDPASREQVLDALRSYTGAVVLVTHDPGAAEALDPQRVVLLPD
MLBr_01816|M.leprae_Br4923 LDEPTNNLDPASRKQVLDALRNYQGAVVLVTHDPGAAKALDPQRVVLLPD
Mb1508|M.bovis_AF2122/97 LDEPTNNLDPASREQVLDALRSYRGAVVLVTHDPGAAAALGPQRVVLLPD
Rv1473|M.tuberculosis_H37Rv LDEPTNNLDPASREQVLDALRSYRGAVVLVTHDPGAAAALGPQRVVLLPD
Mflv_3668|M.gilvum_PYR-GCK LDEPTNNLDPASREQVLDALRSYAGAVVLVTHDPGAAEALDPQRVLLLPD
Mvan_2742|M.vanbaalenii_PYR-1 LDEPTNNLDPASREQVLDALRSYVGSVVLVTHDPGAAEALDPQRVLLLPD
MSMEG_3140|M.smegmatis_MC2_155 LDEPTNNLDPASREQVLDALRSYEGAVVLVTHDPGAAEALDPQRVVLLPD
TH_3270|M.thermoresistible__bu LDEPTNNLDPASREQVLDALRSYVGAVVLVTHDPGAAEALDPQRVVLLPD
MAB_2736c|M.abscessus_ATCC_199 LDEPTNNLDPASREQVLDALRSYLGAVVLVTHDPGAAEALNPQRVVLLPD
*************:*******.* *:*********** **.****:****
MMAR_2279|M.marinum_M ATEDYWSDEYRDLIELA
MUL_1481|M.ulcerans_Agy99 ATEDYWSDEYRDLIELA
MAV_3306|M.avium_104 GTEDYWSDEYRDLIELA
MLBr_01816|M.leprae_Br4923 GTEDYWSDEYRDLIELA
Mb1508|M.bovis_AF2122/97 GTEDYWSDEYRDLIELA
Rv1473|M.tuberculosis_H37Rv GTEDYWSDEYRDLIELA
Mflv_3668|M.gilvum_PYR-GCK GTEDFWSDDYRDLIELA
Mvan_2742|M.vanbaalenii_PYR-1 GTEDFWSDEYRDLIELA
MSMEG_3140|M.smegmatis_MC2_155 GTEDFWSDEYRDLIELA
TH_3270|M.thermoresistible__bu GTEDFWSDEYRDLIELA
MAB_2736c|M.abscessus_ATCC_199 GTEDYWSDEYRDLIELA
.***:***:********