For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRPAIAEPTVNIGEAAALYDVAPSTLRWWEKQGVLSSPAQHSGRRTYTERDLRCIGIAYLCCITGMMPLR QAAIVTSHSAQPDTWRSAVTGQVAEISARIERLHRARDYLNHLLCCQNEDIVDCPYLDGELRLRTPRGRA PGANLISAARSAGYESTPARDETAAGQPGRCGFCGGQLSSHAKGRPQRYCSPACKQRAYRHRHGMK*
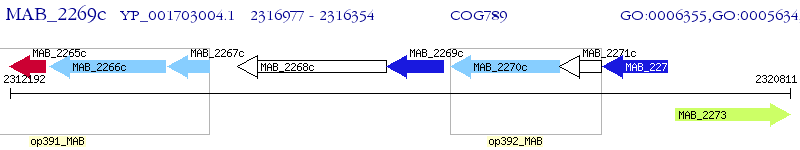
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2269c | - | - | 100% (207) | MerR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3222 | - | 2e-07 | 27.73% (119) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_3180 | - | 8e-10 | 32.17% (115) | transcriptional regulator, MerR family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4691 | - | 6e-07 | 28.23% (124) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3222|M.avium_104 ---------MLTIGEVARRSGVAATTLRYYEQIGLLPAPTRLGGQRRYDE
MSMEG_3180|M.smegmatis_MC2_155 ------MAAHLTIGEVARRAGVAPTALRYYEKLGLLPAPERIGGRRRYDE
Mvan_4691|M.vanbaalenii_PYR-1 ---------MIDISAVARRSGLPTSTLRFYEEKGLIASVGRRSRRRLFGP
MAB_2269c|M.abscessus_ATCC_199 MSRPAIAEPTVNIGEAAALYDVAPSTLRWWEKQGVLSSPAQHSGRRTYTE
: *. .* .:..::**::*: *::.: : . :* :
MAV_3222|M.avium_104 AVLSRLEVIALCKTAGFALDEIQRLFADDAPGRPASRALARAKLAQIDAQ
MSMEG_3180|M.smegmatis_MC2_155 AILVRLEVIRLCKAAGFSLDEIAVLMDDDTPGRPVARALAEAKLAEIDAQ
Mvan_4691|M.vanbaalenii_PYR-1 DVFDRLALITLGRAAGFSLDEIATMLTLGGP-RQIDRKALTTRAAEIDST
MAB_2269c|M.abscessus_ATCC_199 RDLRCIGIAYLCCITGMMPLRQAAIVTSHSAQPDTWRSAVTGQVAEISAR
: : : * :*: . :. . * : *:*.:
MAV_3222|M.avium_104 LDSLTRARAVIEWGMRCTCPS-------------------------IDAC
MSMEG_3180|M.smegmatis_MC2_155 IASLARARSIVEWGMQCTCPS-------------------------IDEC
Mvan_4691|M.vanbaalenii_PYR-1 IERLTAISTALKHAAACPAPDHMECPTFR---------------GYLDLA
MAB_2269c|M.abscessus_ATCC_199 IERLHRARDYLNHLLCCQNEDIVDCPYLDGELRLRTPRGRAPGANLISAA
: * :: * . :. .
MAV_3222|M.avium_104 TCGIHPTRPAPVGPDVRSARRF----------------------------
MSMEG_3180|M.smegmatis_MC2_155 TCGIHRTVPA----------------------------------------
Mvan_4691|M.vanbaalenii_PYR-1 AAGLFSESPPRKSPPKPTIKKQVRLGAGQPIRGE----------------
MAB_2269c|M.abscessus_ATCC_199 RSAGYESTPARDETAAGQPGRCGFCGGQLSSHAKGRPQRYCSPACKQRAY
.. . *.
MAV_3222|M.avium_104 -------
MSMEG_3180|M.smegmatis_MC2_155 -------
Mvan_4691|M.vanbaalenii_PYR-1 -------
MAB_2269c|M.abscessus_ATCC_199 RHRHGMK