For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRSSADDACPGALRLHEAADGALARIRLPGGMLSSAQLAALAEAAGDHGAPILELTSRGNVQLRSVRDAN AVGALLSEAGLLPSPRHERVRNIVASPLSGRAGEFPDARPLVSALDRGLTAAATLAELPGRFLFSLDDGR GDMAAMGADIGLRHDQKGWRLLLGGRTTSVCDGDAVDAVERMLTAARKFVEIRGNAWRVAELPEGAGTLA RAIDAVFIDGPPDPTPAPRAPVGWIEQLRTGESPGELVTLGAAVPLGQLPARVAALLAAVEKPVIVTPWR SVLLCDLPHGDADAVLRVLAPLGLVFDERSPWLNASSCTGLPGCGRSRTDVHADVRLEVAEEIASGIHSP VHRHWVGCPRACGSPSTGQVLVATELGYRPLERHP*
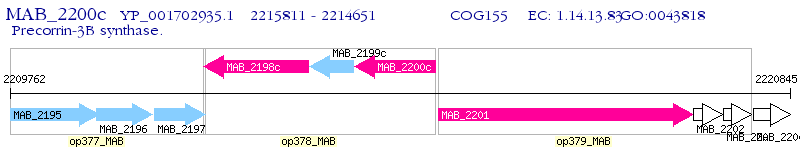
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2200c | - | - | 100% (386) | cobalamin biosynthesis protein CobG |
| M. abscessus ATCC 19977 | MAB_1662c | - | 4e-11 | 23.92% (393) | sulfite reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2090 | cobG | 1e-110 | 57.07% (375) | cobalamin biosynthesis protein CobG |
| M. gilvum PYR-GCK | Mflv_3106 | - | 1e-103 | 54.94% (395) | precorrin-3B synthase |
| M. tuberculosis H37Rv | Rv2064 | cobG | 1e-110 | 57.07% (375) | cobalamin biosynthesis protein CobG |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3044 | - | 1e-105 | 55.06% (385) | cobalamin biosynthesis protein |
| M. avium 104 | MAV_1787 | - | 3e-11 | 24.62% (390) | sulfite reductase |
| M. smegmatis MC2 155 | MSMEG_3871 | cobG | 1e-105 | 54.83% (383) | precorrin-3B synthase |
| M. thermoresistible (build 8) | TH_0438 | cobG | 1e-109 | 56.12% (392) | Possible cobalamin biosynthesis protein CobG |
| M. ulcerans Agy99 | MUL_2288 | - | 1e-104 | 54.81% (385) | cobalamin biosynthesis protein |
| M. vanbaalenii PYR-1 | Mvan_3429 | - | 1e-106 | 55.41% (388) | precorrin-3B synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2090|M.bovis_AF2122/97 ------------------------------------------MAGTRDAD
Rv2064|M.tuberculosis_H37Rv ------------------------------------------MAGTRDAD
MMAR_3044|M.marinum_M ------------------------------------------MTRARDAD
MUL_2288|M.ulcerans_Agy99 ------------------------------------------MTRARDAD
MSMEG_3871|M.smegmatis_MC2_155 ------------------------------------------MTRTRDQD
Mflv_3106|M.gilvum_PYR-GCK ------------------------------------------MVRAREKD
Mvan_3429|M.vanbaalenii_PYR-1 ------------------------------------------MARARDDD
TH_0438|M.thermoresistible__bu ------------------------------------------VERAREHD
MAB_2200c|M.abscessus_ATCC_199 ------------------------------------------MTRSSADD
MAV_1787|M.avium_104 MLAALAIFFRGERPMTTARPAKARNEGQWALGNREPLNPNEEMKQAGAPL
: :
Mb2090|M.bovis_AF2122/97 ACPGALRPHQAADG------------------------------------
Rv2064|M.tuberculosis_H37Rv ACPGALRPHQAADG------------------------------------
MMAR_3044|M.marinum_M ACPGALQVHQAADG------------------------------------
MUL_2288|M.ulcerans_Agy99 ACPGALQVHQAADG------------------------------------
MSMEG_3871|M.smegmatis_MC2_155 ACPGALTVHQAADG------------------------------------
Mflv_3106|M.gilvum_PYR-GCK ACPGALQTHRAADG------------------------------------
Mvan_3429|M.vanbaalenii_PYR-1 ACPGALQTHRAADG------------------------------------
TH_0438|M.thermoresistible__bu ACPGALSVHPAADG------------------------------------
MAB_2200c|M.abscessus_ATCC_199 ACPGALRLHEAADG------------------------------------
MAV_1787|M.avium_104 AVRERIETIYAKNGFDSIDKSDLRGRFRWWGLYTQREQGYDGSWTGDENI
* : * :*
Mb2090|M.bovis_AF2122/97 -------ALARIRLPGGMITAAQLATLASVASDFGSATLELTARGNVQLR
Rv2064|M.tuberculosis_H37Rv -------ALARIRLPGGMITAAQLATLASVASDFGSATLELTARGNVQLR
MMAR_3044|M.marinum_M -------ALVRVRLPGGRLTADQLAALARASNELASATLELTARGNLQLR
MUL_2288|M.ulcerans_Agy99 -------ALVRVRLPGGRLTADQLAALARASNELASATLELTARGNLQLR
MSMEG_3871|M.smegmatis_MC2_155 -------ALVRIRLAGGAITAAQLATLADVANDLGSPAMELTSRGNIQIR
Mflv_3106|M.gilvum_PYR-GCK -------DLSRIRLPGGAITPPQLEVVAQAAIDLAASTLELTSRGNLQIR
Mvan_3429|M.vanbaalenii_PYR-1 -------DLARIRLPGGAITARQLEAVAQAAVNFGTATLELTSRANLQIR
TH_0438|M.thermoresistible__bu -------ALARIRLPGGMLTPAQLTALADAARRFGSEAMELTSRGNIQIR
MAB_2200c|M.abscessus_ATCC_199 -------ALARIRLPGGMLSSAQLAALAEAAGDHGAPILELTSRGNVQLR
MAV_1787|M.avium_104 EKLEARYFMMRVRCDGGAISAAALRTLGQISVDFARDTADITDRENIQYH
: *:* ** ::. * .:. : . ::* * *:* :
Mb2090|M.bovis_AF2122/97 --GIRDVAAVADAVAKAGLLPSATH-ERVRNIVASPLSGR-AGGLADVRA
Rv2064|M.tuberculosis_H37Rv --GIRDVAAVADAVAKAGLLPSATH-ERVRNIVASPLSGR-AGGLADVRA
MMAR_3044|M.marinum_M --AITDVMAVADAIANAGLLPSPTH-ERVRNIVASPLSGR-VGGLADVQS
MUL_2288|M.ulcerans_Agy99 --AITDVMAVADAIANAGLLPSPTH-ERVRNIVASPLSGR-VGGLADLQS
MSMEG_3871|M.smegmatis_MC2_155 --GLTDTDAAAAALAGAGLLPSPTH-ERVRNIVASPLTGR-SGGVADVGA
Mflv_3106|M.gilvum_PYR-GCK --GVRDTAAVADLLAGAGLLPSATH-ERVRNIVASPLSGR-HGGLCDVRA
Mvan_3429|M.vanbaalenii_PYR-1 --GIHDTGAVAALLTDAGLLPSATH-ERVRNIVASPLSGR-SGGFCDARD
TH_0438|M.thermoresistible__bu --GISDPAAVADAVAAAGLLPSPTH-ETVRNIVASPLSGR-CGGATDIRP
MAB_2200c|M.abscessus_ATCC_199 --SVRDANAVGALLSEAGLLPSPRH-ERVRNIVASPLSGR-AGEFPDARP
MAV_1787|M.avium_104 WIEVENVPEIWRRLDAVGLRTTEACGDCPRVILGSPLAGESLEEVIDPSW
: : : .** .: : * *:.***:*. *
Mb2090|M.bovis_AF2122/97 WVGELDAAIRAEPRLAELGGRFWFGLD--DGRADVSGLGADVGVQVFPDG
Rv2064|M.tuberculosis_H37Rv WVGELDAAIRAEPRLAELGGRFWFGLD--DGRADVSGLGADVGVQVFPDG
MMAR_3044|M.marinum_M WVGELDAAICAEPLLAQLGGRFWFSID--DGRADVSGLGADVGVHVLADG
MUL_2288|M.ulcerans_Agy99 WVGELDAAICAEPLLAQLGGRFWFSID--DGRADVSGLGADVGVHVLADG
MSMEG_3871|M.smegmatis_MC2_155 LVSELDAAIQAEPELAGLPGRFLFGLD--DGRGDISGLGPDAGVHMLDAG
Mflv_3106|M.gilvum_PYR-GCK LVADLDAAVQRDPVLAELPGRFLFGFD--DGRGDISGLGADAGVHALDDS
Mvan_3429|M.vanbaalenii_PYR-1 LVRSLDHAVQQDPRLARLPGRFLFGID--DGRGDISGLGADAGIHVLDGH
TH_0438|M.thermoresistible__bu WVTRLDHAIQSELALAALPGRFLFGID--DGRGDIAALAADVAVQTLGAS
MAB_2200c|M.abscessus_ATCC_199 LVSALDRGLTAAATLAELPGRFLFSLD--DGRGDMAAMGADIGLRHDQKG
MAV_1787|M.avium_104 AIAEIARRYIGQPEFADLPRKYKTAISGLQDVAHEVNDVAFIGVNHPEHG
: : :* * :: .:. :. .. . .:.
Mb2090|M.bovis_AF2122/97 P---RLLLTGRDTGVRVA---DVAETLIEVALRFVKIR------------
Rv2064|M.tuberculosis_H37Rv P---RLLLTGRDTGVRVA---DVAETLIEVALRFVKIR------------
MMAR_3044|M.marinum_M C---AVLLDGRDTGVRLAPD-DVVPTLTELAVRFTQIR------------
MUL_2288|M.ulcerans_Agy99 C---AVLLDGRDTGVRLAPD-DVVPTLTELAVRFTQMR------------
MSMEG_3871|M.smegmatis_MC2_155 TG--ALLLGGCDTGVRIDRA-GVVPTLITVARRFAKVH------------
Mflv_3106|M.gilvum_PYR-GCK TA--ALLLAGRDTGVRVALS-SAVDELLAVARRFVDGR------------
Mvan_3429|M.vanbaalenii_PYR-1 TA--ALLLAGRDTGVRLPLG-HVVDELLSVAARFLQVR------------
TH_0438|M.thermoresistible__bu GDEAAVLLAGRDSGVRVAAG-DAVEVMVTVARRFTASR------------
MAB_2200c|M.abscessus_ATCC_199 WR---LLLGGRTTSVCDGDAVDAVERMLTAARKFVEIR------------
MAV_1787|M.avium_104 PGLDLWVGGGLSTNPMLAQRVGAWVPLHEVPEVWAAVTSVFRDYGYRRLR
: * :. . : . :
Mb2090|M.bovis_AF2122/97 --------ETAWRVTELAD-IGELQSGVEL----GPSVRPVT-KTPVGWI
Rv2064|M.tuberculosis_H37Rv --------ETAWRVTELAD-IGELQSGVEL----GPSVRPVT-KTPVGWI
MMAR_3044|M.marinum_M --------GNCWRVNELID-PSVLVPGAAP----GSRFPAVT-KAPVGWI
MUL_2288|M.ulcerans_Agy99 --------GNFWRVNELID-PSVLVPGAAP----GSRFPAVT-KAPVGWI
MSMEG_3871|M.smegmatis_MC2_155 --------GKAWRVGELDD-LAPLLEGLTATGAGGQRWAPTV-RPPVGWI
Mflv_3106|M.gilvum_PYR-GCK --------GKHWRVGELDD-PAILLDGRAPTERPGGTWQART-RPPVGWI
Mvan_3429|M.vanbaalenii_PYR-1 --------GTAWRVDELDD-PTTLLGDRTPTAEPGATWPAHS-RPPVGWI
TH_0438|M.thermoresistible__bu --------GGAWRIAELDD-TSALLDGFARRAAPAPATPAP--RPPIGWI
MAB_2200c|M.abscessus_ATCC_199 --------GNAWRVAELPEGAGTLARAIDAVFIDGPPDPTPAPRAPVGWI
MAV_1787|M.avium_104 SKARLKFLVKDWGIEKFREVLETEYLKRPLIDGPAPEPVAHP-IDHVGVQ
* : :: : . . :*
Mb2090|M.bovis_AF2122/97 PQDDSR------VTLGAAVPLGVLPARVAECLAAIEAPLVITPWRSVLIC
Rv2064|M.tuberculosis_H37Rv PQDDSR------VTLGAAVPLGVLPARVAECLAAIEAPLVITPWRSVLIC
MMAR_3044|M.marinum_M PQRDGR------VTLGAAVPLGILPARVAEFLAAIEAPIAITPWRSVLVF
MUL_2288|M.ulcerans_Agy99 PQRDGR------VTLGAAVPLGILPARVAEFLAAIEAPIAITPWRSVLVF
MSMEG_3871|M.smegmatis_MC2_155 PQRDGR------VALGAGVPLGVLSARDAQFLAAIEAPIVITPWRSVLVF
Mflv_3106|M.gilvum_PYR-GCK EQTDGR------IALGAAVPLGVLDAQVARYLAAVEAPLTVTPWRSVLIF
Mvan_3429|M.vanbaalenii_PYR-1 EQDDGR------IALGAAVPLGVLDAQVARYLAAVEAPLAVTPWRSVLLF
TH_0438|M.thermoresistible__bu PQDDGR------VTLAAGVPLGVLTARLAEFLAAIDAPLIVTPWRSVLVC
MAB_2200c|M.abscessus_ATCC_199 EQLRTGESPGELVTLGAAVPLGQLPARVAALLAAVEKPVIVTPWRSVLLC
MAV_1787|M.avium_104 RLKNGLNAVG-VAPIAGRVSGTILLAVADLAQQAGCDRIRFTPYQKLVLL
.:.. *. * * * : .**::.:::
Mb2090|M.bovis_AF2122/97 DLDDATADAALRVLAPLGLVFDENSPWLNISACTGSPGCAHSAADVRADA
Rv2064|M.tuberculosis_H37Rv DLDDATADAALRVLAPLGLVFDENSPWLNISACTGSPGCAHSAADVRADA
MMAR_3044|M.marinum_M DLDEAIADTALRVLAPLGLVFDENSPWLNVSACTGSPGCASSAADVRADA
MUL_2288|M.ulcerans_Agy99 DLDEAIADTALRVLAPLGLVFDENSPWLNVSACTGSPGCASSAADVRADA
MSMEG_3871|M.smegmatis_MC2_155 DLDEGVADTALRVLAPMGLVFDENSPWLDVSACTGSPGCAKSLTDVRADA
Mflv_3106|M.gilvum_PYR-GCK DLDEGVADVALRVLAPLGLIFDEDSAWLSVSACTGSPGCAHSAADVRADA
Mvan_3429|M.vanbaalenii_PYR-1 DLDEGVADVALRVLAPLGLVFDENSPWLSVSACTGSPGCERSVADVRADA
TH_0438|M.thermoresistible__bu DLDEGVADTAVRVLAPLGLIFDENSPWLRISSCTGLPGCARSAADVRADA
MAB_2200c|M.abscessus_ATCC_199 DLPHGDADAVLRVLAPLGLVFDERSPWLNASSCTGLPGCGRSRTDVHADV
MAV_1787|M.avium_104 DIPDDKLDEVVAGLEALGLQSQPSHWRRNLMACSGIEFCKLSFAETRVRA
*: . * .: * .:** : :*:* * * ::.:. .
Mb2090|M.bovis_AF2122/97 ARSLNVESAG------------HRHFVGCERACGSPPAGEV-----LVAT
Rv2064|M.tuberculosis_H37Rv ARSLNVESAG------------HRHFVGCERACGSPPAGEV-----LVAT
MMAR_3044|M.marinum_M ALSVRTEAAASGV---------HRHFVGCERACGSPLAGEV-----LVAT
MUL_2288|M.ulcerans_Agy99 ALSVRTEAAASGV---------HRHFVGCERACGSPLAGEV-----LVAT
MSMEG_3871|M.smegmatis_MC2_155 AAAAQEPSAN------------HRHFVGCERACGSPPVGDV-----LVAT
Mflv_3106|M.gilvum_PYR-GCK GAAVDPSIAV------------HRHFVGCDRACGSPADAQV-----LVAT
Mvan_3429|M.vanbaalenii_PYR-1 AAAVDDPVTV------------HRHFVGCERACGSPADGEV-----LVAT
TH_0438|M.thermoresistible__bu ARAATEAAPXGQTR-------VHRHFVGCERACGTPPGAQV-----LVAT
MAB_2200c|M.abscessus_ATCC_199 RLEVAEEIASGIHSP------VHRHWVGCPRACGSPSTGQV-----LVAT
MAV_1787|M.avium_104 QGLVPELERRLADVNRQLDVPITINLNGCPNSCARIQVADIGLKGQMVDD
: ** .:*. .:: :*
Mb2090|M.bovis_AF2122/97 GGG--------------------YRRLRP---------------------
Rv2064|M.tuberculosis_H37Rv GGG--------------------YRRLRP---------------------
MMAR_3044|M.marinum_M AQG--------------------YRLLRKA--------------------
MUL_2288|M.ulcerans_Agy99 AQG--------------------YRLLRKA--------------------
MSMEG_3871|M.smegmatis_MC2_155 EDG--------------------YRLRIASP-------------------
Mflv_3106|M.gilvum_PYR-GCK GDG--------------------YRSREAHP-------------------
Mvan_3429|M.vanbaalenii_PYR-1 GDG--------------------YRARKRHP-------------------
TH_0438|M.thermoresistible__bu GEG--------------------YRLRNPHP-------------------
MAB_2200c|M.abscessus_ATCC_199 ELG--------------------YRPLERHP-------------------
MAV_1787|M.avium_104 GEGGSVEGFQVHLGGSLGQDSGFGRKLRQHKVTSDELGDYIERVARNFLK
* *
Mb2090|M.bovis_AF2122/97 ---------------------
Rv2064|M.tuberculosis_H37Rv ---------------------
MMAR_3044|M.marinum_M ---------------------
MUL_2288|M.ulcerans_Agy99 ---------------------
MSMEG_3871|M.smegmatis_MC2_155 ---------------------
Mflv_3106|M.gilvum_PYR-GCK ---------------------
Mvan_3429|M.vanbaalenii_PYR-1 ---------------------
TH_0438|M.thermoresistible__bu ---------------------
MAB_2200c|M.abscessus_ATCC_199 ---------------------
MAV_1787|M.avium_104 YRGEGERFAQWAMRADEDDLR