For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSPAPVALAAGDEMPSSQFGPLTRSHIVRYAGSGGDFNPIHHDEEFARAAGMPGVFGMGLLHGGVLAQ RLTAWVGLANIRSFRIRFTGQVWPGDLLSFGGRVTGVREAGGQQVADLELVVTRQSGEPAIKGSAVVQVA R
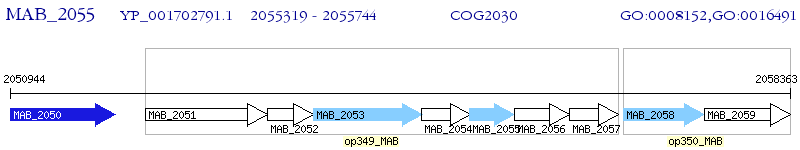
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2055 | - | - | 100% (141) | MaoC-like dehydratase |
| M. abscessus ATCC 19977 | MAB_2057 | - | 5e-13 | 31.06% (132) | hypothetical protein MAB_2057 |
| M. abscessus ATCC 19977 | MAB_4780 | - | 5e-11 | 33.33% (132) | MaoC-like dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0655 | - | 4e-14 | 32.84% (134) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
| M. gilvum PYR-GCK | Mflv_5120 | - | 2e-12 | 33.58% (134) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
| M. tuberculosis H37Rv | Rv0636 | - | 7e-14 | 32.84% (134) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
| M. leprae Br4923 | MLBr_01909 | - | 5e-13 | 36.19% (105) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
| M. marinum M | MMAR_0969 | - | 4e-13 | 33.85% (130) | hypothetical protein MMAR_0969 |
| M. avium 104 | MAV_2649 | - | 2e-30 | 47.24% (127) | MaoC like domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1341 | - | 2e-11 | 36.19% (105) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
| M. thermoresistible (build 8) | TH_1867 | - | 4e-14 | 33.58% (134) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0721 | - | 3e-13 | 33.85% (130) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
| M. vanbaalenii PYR-1 | Mvan_3988 | - | 2e-19 | 37.21% (129) | dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0969|M.marinum_M --------MALREFSSVNVGDQLP-ERTYPLTRQDLVNYAGVSGDLNPIH
MUL_0721|M.ulcerans_Agy99 --------MALREFSSVNVGDQLP-ERTYPLTRQDLVNYAGVSGDLNPIH
Mb0655|M.bovis_AF2122/97 --------MALREFSSVKVGDQLP-EKTYPLTRQDLVNYAGVSGDLNPIH
Rv0636|M.tuberculosis_H37Rv --------MALREFSSVKVGDQLP-EKTYPLTRQDLVNYAGVSGDLNPIH
MLBr_01909|M.leprae_Br4923 --------MARREFSSVKVGDLLP-EKTYPLTRQDLVNYAGVSGDLNPIH
Mflv_5120|M.gilvum_PYR-GCK --------MALREFSSVKVGDLLP-EKVIPLTRADLVNYAGVSGDLNPIH
MSMEG_1341|M.smegmatis_MC2_155 --------MALREFSSVKVGDTLP-ERVITLTRGDLVNYAGVSGDLNPIH
TH_1867|M.thermoresistible__bu --------MALRKFDSVKVGDKLP-EKVIPLTRADLVNYAGVSGDLNPIH
MAB_2055|M.abscessus_ATCC_1997 MT-------TSPAPVALAAGDEMPSSQFGPLTRSHIVRYAGSGGDFNPIH
MAV_2649|M.avium_104 MSPGDGAGGQTTTTTALRVGVQAPVREFGPLTRQMFVRYAGASGDFNPMH
Mvan_3988|M.vanbaalenii_PYR-1 ----------MIGIDSVAVGDELPPLQAPDVSRLTLALFAGASGDHNPMH
:: .* * . ::* :. :** .** **:*
MMAR_0969|M.marinum_M WDDEIAKLVGLDTAIAHGMLTMGIGGGYVTSWVGDPGAVTEYNVRFTAVV
MUL_0721|M.ulcerans_Agy99 WDDEIAKLVGLDTAIAHGMLTMGIGGGYVTSWVGDPGAVTEYNVRFTAVV
Mb0655|M.bovis_AF2122/97 WDDEIAKVVGLDAAIAHGMLTMGIGGGYVTSWVGDPGAVTEYNVRFTAVV
Rv0636|M.tuberculosis_H37Rv WDDEIAKVVGLDTAIAHGMLTMGIGGGYVTSWVGDPGAVTEYNVRFTAVV
MLBr_01909|M.leprae_Br4923 WDDEIAKVVGLDTAIAHGMLTMGIGGGYVTSWVGDPGAVIEYNVRFTAVV
Mflv_5120|M.gilvum_PYR-GCK WDDEIAKQVGLDTAIAHGMLTMGLGGGYVTEWVGDPAAVTEYNVRFTAVV
MSMEG_1341|M.smegmatis_MC2_155 WDDEIAKQVGLDTAIAHGMLTMGLGGGYVTSWVGDPAAVTEYNVRFTAVV
TH_1867|M.thermoresistible__bu WDDEIAKQVGLDTAIAHGMLTMGMGGGYITEWVGDPAAVTEYNVRFTAVV
MAB_2055|M.abscessus_ATCC_1997 HDEEFARAAGMPGVFGMGLLHGGVLAQRLTAWVG-LANIRSFRIRFTGQV
MAV_2649|M.avium_104 YDDQLARSAGYPSVFAQGMFSAALLAGFATDWLG-AGSVRRFGVRFREQV
Mvan_3988|M.vanbaalenii_PYR-1 VDLDEARSAGLPDVFAHGMLSMAYLGRLITDWVP-QERLRTFRVRFAAIT
* : *: .* .:. *:: . . * *: : : :** .
MMAR_0969|M.marinum_M PVPNDGKGAELVFNGRVKSVDPDSKS----VTIALTATTGGKKIFGRAIA
MUL_0721|M.ulcerans_Agy99 PVPNDGKGAELVFNGRVKSVDPDSKS----VTIALTATTGGKKIFGRAIA
Mb0655|M.bovis_AF2122/97 PVPNDGKGAELVFNGRVKSVDPESKS----VTIALTATTGGKKIFGRAIA
Rv0636|M.tuberculosis_H37Rv PVPNDGKGAELVFNGRVKSVDPESKS----VTIALTATTGGKKIFGRAIA
MLBr_01909|M.leprae_Br4923 PVPNDGQGAVLVFSGKVKSVDPDTKS----VTIALSATTGGKKIFGRAIA
Mflv_5120|M.gilvum_PYR-GCK PVPNDGKGAEIVFNGRVKSVDPESRS----VTIALSATSGGKKIFGRAVA
MSMEG_1341|M.smegmatis_MC2_155 PVPNDGVGAEITFNGRVKSVDAEEKL----VTIAISATAGGKKIFGRAVA
TH_1867|M.thermoresistible__bu PVPNDGKGAEIVFGGRVKSVDPETKS----VTIALTATAGGKKIFGRAIA
MAB_2055|M.abscessus_ATCC_1997 WP-----GDLLSFGGRVTGVREAGGQQVADLELVVTRQSGEPAIKGSAVV
MAV_2649|M.avium_104 WP-----GDVLTCSGTVTAVSAEPDAERVIVELTATRQTGGVAITGTAEF
Mvan_3988|M.vanbaalenii_PYR-1 PV-----HARPTCTGRVTDISVVDGERRATVEIVVTLADGTITLSGDAVV
* *. : : :. : * : * *
MMAR_0969|M.marinum_M SAKLA------------
MUL_0721|M.ulcerans_Agy99 SAKLA------------
Mb0655|M.bovis_AF2122/97 SAKLA------------
Rv0636|M.tuberculosis_H37Rv SAKLA------------
MLBr_01909|M.leprae_Br4923 SAKLA------------
Mflv_5120|M.gilvum_PYR-GCK IAKLA------------
MSMEG_1341|M.smegmatis_MC2_155 TARLA------------
TH_1867|M.thermoresistible__bu TAKLA------------
MAB_2055|M.abscessus_ATCC_1997 QVAR-------------
MAV_2649|M.avium_104 VNPRNAARCGAGDADRP
Mvan_3988|M.vanbaalenii_PYR-1 GL---------------