For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRRGRVARITAVLLLATLPLLSGCVRVKASMTVTTDDHVSGQLIAVAKPQGKNDKGPQLDRNMSFAEKVQ VSEYEENGMVGSRAIFNDLTFSEVPQLAGMNKNAAGFDLTLRRNGDVVVLEGRADLTALGDNSNADVSLA VSFPGDIESTNGEMLGSDSVQWKLNPGVVSSFTATARYTDPSTRSFNGAAIFVSIAALLVGALVASMAWL DRDQSPRFTPEQFPE*
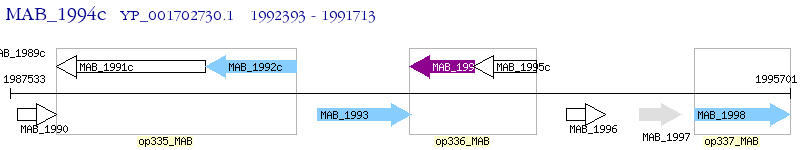
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1994c | - | - | 100% (226) | lipoprotein LppM |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2193 | lppM | 4e-67 | 54.71% (223) | lipoprotein lppM |
| M. gilvum PYR-GCK | Mflv_2976 | - | 3e-67 | 58.06% (217) | LppM |
| M. tuberculosis H37Rv | Rv2171 | lppM | 4e-67 | 54.71% (223) | lipoprotein lppM |
| M. leprae Br4923 | MLBr_00902 | - | 3e-61 | 51.61% (217) | putative lipoprotein |
| M. marinum M | MMAR_3206 | lppM | 2e-65 | 54.34% (219) | lipoprotein LppM |
| M. avium 104 | MAV_2323 | - | 4e-65 | 54.79% (219) | LppM protein |
| M. smegmatis MC2 155 | MSMEG_4239 | - | 3e-66 | 56.68% (217) | LppM protein |
| M. thermoresistible (build 8) | TH_0707 | lppM | 3e-63 | 56.46% (209) | Probable conserved lipoprotein lppM |
| M. ulcerans Agy99 | MUL_3514 | lppM | 2e-64 | 55.29% (208) | lipoprotein LppM |
| M. vanbaalenii PYR-1 | Mvan_3535 | - | 2e-67 | 57.75% (213) | LppM |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2976|M.gilvum_PYR-GCK ----------MLTR--HRTRRRLLAMVLLLLVVAPSLIGCVRVRASITVS
Mvan_3535|M.vanbaalenii_PYR-1 ----------MLNR--HRARRRLFALVLLLLVVAPSLIGCVRIRASITVS
MSMEG_4239|M.smegmatis_MC2_155 ----------MRVRRPSRSRRRLLVLVMLLLLVLPTAVGCVRIRASITVS
TH_0707|M.thermoresistible__bu -----------------------VIAVLALLLMLPALVGCVRVRASITVS
Mb2193|M.bovis_AF2122/97 --------------MARTRRRGMLAIAMLLM-LVPLATGCLRVRASITIS
Rv2171|M.tuberculosis_H37Rv --------------MARTRRRGMLAIAMLLM-LVPLATGCLRVRASITIS
MMAR_3206|M.marinum_M MRASSPPLSAPAAPALRARRRRLLAIAMLLM-LVPLATGCLRVRASITIS
MUL_3514|M.ulcerans_Agy99 ---------------------------MLLM-LVPLATGCLRVRASITIS
MAV_2323|M.avium_104 -----------------MRRRRLLAFAMLLL-VVPLATGCLRVRASLTIS
MLBr_00902|M.leprae_Br4923 MRARFPPLFTRG---CTAQRRRTLTIALLLVAMVPLATGCLRVTASITIS
MAB_1994c|M.abscessus_ATCC_199 ---------------MTRRGRVARITAVLLLATLPLLSGCVRVKASMTVT
: *: * **:*: **:*::
Mflv_2976|M.gilvum_PYR-GCK PDDRVSGQIVAAAIPRDDNDKGPQLLNN-LPFAQKVAVSDYSRDDYVGSQ
Mvan_3535|M.vanbaalenii_PYR-1 PDDRVSGQIVAAAIPRDDNDKGPQLLNN-LPFAQKVAVSDYSRDDYVGSQ
MSMEG_4239|M.smegmatis_MC2_155 PDDRVSGQIIAAAKPSDADDKGPQLVNN-LPFAHKVAVSEYSRDDYVGSQ
TH_0707|M.thermoresistible__bu PDDRVSGQIIAAAKVEDPADEGPQLVNS-LPFANKVSVSRYERDDYVGTQ
Mb2193|M.bovis_AF2122/97 PDDLVSGEIIAAAKPKNSKDTGPALDGD-VPFSQKVAVSNYDSDGYVGSQ
Rv2171|M.tuberculosis_H37Rv PDDLVSGEIIAAAKPKNSKDTGPALDGD-VPFSQKVAVSNYDSDGYVGSQ
MMAR_3206|M.marinum_M PEDLVSGEIIAAAKPKNNKDTGPQLDSDNVAFSQKVAVSNYDSDGYVGSQ
MUL_3514|M.ulcerans_Agy99 PEDLVSGEIIAAAKPKNNKDTGPQLDSDNVAFSQKVAVSNYDGDGYVGSQ
MAV_2323|M.avium_104 PDDLVSGEIVAAAKPKTPKDTGPQLDSNNLPFSQKVAVSNYDSDGYVGSQ
MLBr_00902|M.leprae_Br4923 PDNLVSGKIIAAAKPKNKNDAGPQLNDN-LPFSQKIAVSNYNSDGYVGSQ
MAB_1994c|M.abscessus_ATCC_199 TDDHVSGQLIAVAKPQGKNDKGPQLDRN-MSFAEKVQVSEYEENGMVGSR
.:: ***:::*.* * ** * . :.*:.*: ** *. :. **::
Mflv_2976|M.gilvum_PYR-GCK ATFSDLTFAELPQLAAMNRDAAGVDISLRRAGDLVILEGRVDLTSLSD-P
Mvan_3535|M.vanbaalenii_PYR-1 AVFSDLTFAELPQLASMNRDAAGVDISLRRAGDLVILEGRADLTSLSD-P
MSMEG_4239|M.smegmatis_MC2_155 AVFSDLSFAEVPQLAALSRDAAGVDISLRRAGDLVILEGRVDLTSMND-P
TH_0707|M.thermoresistible__bu ATFSGLTFAELPQLANLNREAAGMDLSLRRAGNVVILEGRVDLTTLND-P
Mb2193|M.bovis_AF2122/97 AVFSDLTFAELPQLANMNSDAAGVNLSLRRNGNIVILEGRADLTSVSD-P
Rv2171|M.tuberculosis_H37Rv AVFSDLTFAELPQLANMNSDAAGVNLSLRRNGNIVILEGRADLTSVSD-P
MMAR_3206|M.marinum_M AVFSDLTFAELPQLANMNPDAAGVNLSLRRNGNMVILEGRVDLTSVTD-A
MUL_3514|M.ulcerans_Agy99 AVFSDLTFAELPQLANMNPDAAGVNLSLRRNGNMVILEGRVDLTSVTD-A
MAV_2323|M.avium_104 AVFSDLTFAELPQLANMNSDAAGVNLSLRRNGNLVLLEGRADLTSLTD-P
MLBr_00902|M.leprae_Br4923 AVFSDLTFAELPQLANMNSSTTDVTLSLRRNGNLVILESRADLTSVTD-P
MAB_1994c|M.abscessus_ATCC_199 AIFNDLTFSEVPQLAGMNKNAAGFDLTLRRNGDVVVLEGRADLTALGDNS
* *..*:*:*:**** :. .::.. ::*** *::*:**.*.***:: * .
Mflv_2976|M.gilvum_PYR-GCK EADVSLSVAFPGEVTSTNGDQISSSVVEWKLRPGVVSTMNAQARYTDPSA
Mvan_3535|M.vanbaalenii_PYR-1 EADVSLSVAFPGEVTSTNGEQISSSVVEWKLRPGVVSTMNAQARYTDPSA
MSMEG_4239|M.smegmatis_MC2_155 EADVLLTVSFPGDVTSTNGDQVTSSIVEWKLRPGVVSTMNAQARYTDPSA
TH_0707|M.thermoresistible__bu QADVSLTVSFPGEVTSTNGNRISAEIVEWKLRPGVVSTMSAQARYTDPST
Mb2193|M.bovis_AF2122/97 DADVELTVAFPAAVTSTNGDRIEPEVVQWKLKPGVVSTMSAQARYTDPNT
Rv2171|M.tuberculosis_H37Rv DADVELTVAFPAAVTSTNGDRIEPEVVQWKLKPGVVSTMSAQARYTDPNT
MMAR_3206|M.marinum_M DAEVELTVAFPGTVTSTNGDRIEPEVVQWKLKPGVVSTMNAQARYTDPNT
MUL_3514|M.ulcerans_Agy99 DAEVELTVAFPGTVTSTNGDRIEPEVVQWKLKPGVVSTMNAQARYTDPNT
MAV_2323|M.avium_104 EADVELTVAFPGVVTSTNGDRVEPDVVSWKLKPGVVSTMTAQARYTDPNT
MLBr_00902|M.leprae_Br4923 DADVELTVAFPGVVTSTNGDRIETKVVAWKLKPGVVSTMSARARYTDPDT
MAB_1994c|M.abscessus_ATCC_199 NADVSLAVSFPGDIESTNGEMLGSDSVQWKLNPGVVSSFTATARYTDPST
:*:* *:*:**. : ****: : .. * ***.*****::.* ******.:
Mflv_2976|M.gilvum_PYR-GCK RSFAGAAIWLGVASFLVAGIVAWLAYSNRDRSPRPGEPQEQTH--
Mvan_3535|M.vanbaalenii_PYR-1 RSFTGAAIWLGMASLVVAGIIGWLAYSNRDRSPRPGETQEPTR--
MSMEG_4239|M.smegmatis_MC2_155 RSFTTAAIWLVIAAFVVASVVAGLAYVSRDQSPKPGEPAPTET--
TH_0707|M.thermoresistible__bu RSFVKAAIWLGVVSFAVAAVIAVLAWRSRDRSPKVWEIQPQDS--
Mb2193|M.bovis_AF2122/97 RSFTGAGIWLGIAAFAAAGVVAVLAWIDRDRSPRLTASGDPPTS-
Rv2171|M.tuberculosis_H37Rv RSFTGAGIWLGIAAFAAAGVVAVLAWIDRDRSPRLTASGDPPTS-
MMAR_3206|M.marinum_M RSFTAAGIWLAIASFAAAGVVAALAWVSRDRSPRFTAPGDQPPS-
MUL_3514|M.ulcerans_Agy99 RSFTAAGIWLAIASFAAAGAVAALAWVSRDRSPRFTAPGDQPPS-
MAV_2323|M.avium_104 RSFTGAVVWLGIASFAAAGVVGLLAWVSRDRSPRLTAPRDQPPPE
MLBr_00902|M.leprae_Br4923 RSFTGAAVWLGIASFSAASVVVLLAWNERKSSARLQIPRDSSSS-
MAB_1994c|M.abscessus_ATCC_199 RSFNGAAIFVSIAALLVGALVASMAWLDRDQSPRFTPEQFPE---
*** * ::: :.:: ... : :*: .*. *.: