For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTEIGTEAVLNQTETLTDLESESIHIIREVAGEFERPVLLFSGGKDSTVLLHIALKAFWPAPLPFSLLHV DTGHNLPEVLAFRDEIVERHNLRLVVSNVEDYLADGRLTERPDGIRNPLQTIPLLEAITDNKFDAVLGGG RRDEERSRAKERIFSLRNAFGQWDPKKQRPELWSLYNGRHAPGEHVRVFPISNWTELDVWRYIARENVQL ASLYYAHEREVFNRDGMLLTPGPWGGPRDGEELQTLSVRYRTVGDGSTTGAVLSDAADVHAVLAEVASSR ITERGATRGDDRVSEAAMEDRKREGYF*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
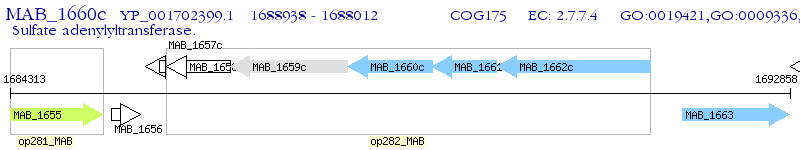
| M. abscessus ATCC 19977 | MAB_1660c | - | - | 100% (308) | sulfate adenylyltransferase subunit 2 |
| M. abscessus ATCC 19977 | MAB_4181 | - | e-107 | 62.24% (294) | sulfate adenylyltransferase subunit 2 |
| M. abscessus ATCC 19977 | MAB_1545c | - | 6e-98 | 58.92% (297) | sulfate adenylyltransferase subunit 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1316 | cysD | 1e-104 | 62.16% (296) | sulfate adenylyltransferase subunit 2 |
| M. gilvum PYR-GCK | Mflv_2271 | - | 1e-106 | 61.89% (307) | sulfate adenylyltransferase subunit 2 |
| M. tuberculosis H37Rv | Rv1285 | cysD | 1e-104 | 62.16% (296) | sulfate adenylyltransferase subunit 2 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4133 | cysD | 1e-104 | 59.55% (309) | sulfate adenylyltransferase subunit 2 CysD |
| M. avium 104 | MAV_1436 | cysD | 1e-110 | 62.58% (310) | sulfate adenylyltransferase subunit 2 |
| M. smegmatis MC2 155 | MSMEG_4979 | cysD | 1e-111 | 65.41% (292) | sulfate adenylyltransferase subunit 2 |
| M. thermoresistible (build 8) | TH_2243 | cysD | 1e-109 | 65.41% (292) | PROBABLE SULFATE ADENYLYLTRANSFERASE SUBUNIT 2 CYSD |
| M. ulcerans Agy99 | MUL_3996 | cysD | 1e-103 | 59.22% (309) | sulfate adenylyltransferase subunit 2 |
| M. vanbaalenii PYR-1 | Mvan_4423 | - | 1e-107 | 62.21% (307) | sulfate adenylyltransferase subunit 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1316|M.bovis_AF2122/97 MAITINMVNPTGFIRYEDVEQEAMTSDVTVGPAPGQYQLSHLRLLEAEAI
Rv1285|M.tuberculosis_H37Rv MAITINMVNPTGFIRYEDVEQEAMTSDVTVGPAPGQYQLSHLRLLEAEAI
MMAR_4133|M.marinum_M ----------------------MTTSGVTTGPAVGQYELSHLRSLEAEAM
MUL_3996|M.ulcerans_Agy99 ----------------------MTTSGVTTGPAVGQYELSHLRSLEAEAM
MAV_1436|M.avium_104 ------------------MPKEAMTADLQTAPAAGQYELSHLRSLEAEAI
Mflv_2271|M.gilvum_PYR-GCK -----------------------MTATDELTHNAGRYELSHLRALEAEAI
Mvan_4423|M.vanbaalenii_PYR-1 -----------------------MTATDELTQNAGRYELSHLRALEAEAI
TH_2243|M.thermoresistible__bu --------------MTATIPQASEGAPGSGAPEPRAYELSHLRALEAEAI
MSMEG_4979|M.smegmatis_MC2_155 ---------------------MTAVDSAPAQTRTGQYELSHLRALEAEAI
MAB_1660c|M.abscessus_ATCC_199 -------------------------MTTEIGTEAVLNQTETLTDLESESI
: . * **:*::
Mb1316|M.bovis_AF2122/97 HVIREVAAEFERPVLLFSGGKDSIVMLHLALKAFRPGRLPFPVMHVDTGH
Rv1285|M.tuberculosis_H37Rv HVIREVAAEFERPVLLFSGGKDSIVMLHLALKAFRPGRLPFPVMHVDTGH
MMAR_4133|M.marinum_M HIIREVAAEFERPVLLFSGGKDSIVMLHLAIKAFRPGRLPFPVMHVDTGH
MUL_3996|M.ulcerans_Agy99 HIIREVAAEFERPVLLFSGGKDSIVMLHLAIKAFRPGRLPFPVMHVDTGH
MAV_1436|M.avium_104 HIIREVAAEFERPVLLFSGGKDSIVMLHLALRAFRPGRLPFPVMHVDTGH
Mflv_2271|M.gilvum_PYR-GCK HIIREVAAEFERPVLLFSGGKDSIVMLHLAVKAFRPGRLPFPVMHVDTGH
Mvan_4423|M.vanbaalenii_PYR-1 HIIREVAAEFERPVLLFSGGKDSIVMLHLAIKAFRPGRLPFPVMHVDTGH
TH_2243|M.thermoresistible__bu HIIREVAAEFERPVLLFSGGKDSIVMLHLAMKAFRPGRVPFPVMHVDTGH
MSMEG_4979|M.smegmatis_MC2_155 HIIREVAAEFERPVLLFSGGKDSIVMLHLAIKAFAPGRLPFPVMHVDTGH
MAB_1660c|M.abscessus_ATCC_199 HIIREVAGEFERPVLLFSGGKDSTVLLHIALKAFWPAPLPFSLLHVDTGH
*:*****.*************** *:**:*::** *. :**.::******
Mb1316|M.bovis_AF2122/97 NFDEVIATRDELVAAAGVRLVVASVQDDIDAGRVVETIP-SRNPIQTVTL
Rv1285|M.tuberculosis_H37Rv NFDEVIATRDELVAAAGVRLVVASVQDDIDAGRVVETIP-SRNPIQTVTL
MMAR_4133|M.marinum_M NFDEVIATRDELVAASGVRLVVASVQDDIDAGRVVETFP-SRNPMQTVTL
MUL_3996|M.ulcerans_Agy99 NFDEVIATRDELVAASGVRLVVASVQDDIDAGRVVETFP-SRNSMQTVTL
MAV_1436|M.avium_104 NFDEVIATRDELVKQHGLRLVVASVQDDIDAGRVVEPKQ-GRNPIQTVTL
Mflv_2271|M.gilvum_PYR-GCK NFDEVLQARDELVAETGVRLVVAKVQDDIDAGRVVETIP-SRNPMQTFTL
Mvan_4423|M.vanbaalenii_PYR-1 NFEEVLAARDELVAESGVRLVVAKVQDDIDAGRVVETIP-SRNPMQTFTL
TH_2243|M.thermoresistible__bu NFDEVLATRDELVAENNLRLVVAKVQDDIDAGRVVENGP-SRNPLQTVTL
MSMEG_4979|M.smegmatis_MC2_155 NFEEVIATRDALVEKHGLRLVVASVQDDIDAGRVVDNGP-SRNPLQTVTL
MAB_1660c|M.abscessus_ATCC_199 NLPEVLAFRDEIVERHNLRLVVSNVEDYLADGRLTERPDGIRNPLQTIPL
*: **: ** :* .:****:.*:* : **:.: **.:**..*
Mb1316|M.bovis_AF2122/97 LRAIRENQFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKAQRPELWN
Rv1285|M.tuberculosis_H37Rv LRAIRENQFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKAQRPELWN
MMAR_4133|M.marinum_M LRAIRENRFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKAQRPELWN
MUL_3996|M.ulcerans_Agy99 LRAIRENRFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKAQRPELWN
MAV_1436|M.avium_104 LRAIRENKFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKAQRPELWN
Mflv_2271|M.gilvum_PYR-GCK LRAIRENKFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKNQRPELWN
Mvan_4423|M.vanbaalenii_PYR-1 LRAIRENKFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKNQRPELWN
TH_2243|M.thermoresistible__bu LRAIRENKFDAAFGGARRDEEKARAKERVFSFRDEFGQWNPKAQRPELWN
MSMEG_4979|M.smegmatis_MC2_155 LRGIRENKFDAAFGGARRDEEKARAKERVFSFRDEFGQWDPKAQRPELWN
MAB_1660c|M.abscessus_ATCC_199 LEAITDNKFDAVLGGGRRDEERSRAKERIFSLRNAFGQWDPKKQRPELWS
*..* :*:***.:**.*****::*****:**:*: ****:** ******.
Mb1316|M.bovis_AF2122/97 LYNGRHHKGEHIRVFPLSNWTEFDIWSYIGAEQVRLPSIYFAHRRKVFQR
Rv1285|M.tuberculosis_H37Rv LYNGRHHKGEHIRVFPLSNWTEFDIWSYIGAEQVRLPSIYFAHRRKVFQR
MMAR_4133|M.marinum_M LYNGRHHKGEHIRVFPLSNWTEFDIWSYIGAEHITLPSIYFAHRRKVFKR
MUL_3996|M.ulcerans_Agy99 LYNGRHHKGEHIRVFPLSNWTEFDIWSYIGAEHITLPSIYFAHRRKVFKR
MAV_1436|M.avium_104 LYNGRHHKGEHIRVFPLSNWTEFDIWSYIGAENIALPSIYYAHRRKVFRR
Mflv_2271|M.gilvum_PYR-GCK LYNGRHRKGEHIRAFPLSNWTEFDIWSYIGAEQIKLPSIYYAHERKVFER
Mvan_4423|M.vanbaalenii_PYR-1 LYNGRHRKGEHIRAFPLSNWTEFDIWSYIGAEKIKLPSIYYAHQRKVFER
TH_2243|M.thermoresistible__bu LYNGRHHKGEHIRVFPLSNWTEFDIWSYIGAENITLPSIYYAHRRKVFRR
MSMEG_4979|M.smegmatis_MC2_155 IYNGRHRKGEHIRVFPLSNWTEYDIWAYIGAEDITLPSIYYAHQRKVFGR
MAB_1660c|M.abscessus_ATCC_199 LYNGRHAPGEHVRVFPISNWTELDVWRYIARENVQLASLYYAHEREVFNR
:***** ***:*.**:***** *:* **. *.: *.*:*:**.*:** *
Mb1316|M.bovis_AF2122/97 DGMLLAVHRHMQPRADEPVFEATVRFRTVGDVTCTGCVESSASTVAEVIA
Rv1285|M.tuberculosis_H37Rv DGMLLAVHRHMQPRADEPVFEATVRFRTVGDVTCTGCVESSASTVAEVIA
MMAR_4133|M.marinum_M DGMLLAVDRHMQPGADEPVFEATVRFRTVGDVTCTGCVESEAATVAEVIA
MUL_3996|M.ulcerans_Agy99 DGMLLAVDRHMQPGADEPVFEATVRFRTVGDVTCTGCVESEAATVAEVIA
MAV_1436|M.avium_104 DGMLLAVDRNMQPNPDEPVFEATVRFRTVGDVTCTGCVESQAATVDEVIA
Mflv_2271|M.gilvum_PYR-GCK DGMLLAVHKYLQPRKDEPIIEKTVRFRTVGDVTCTGCVESTAATVSEVIA
Mvan_4423|M.vanbaalenii_PYR-1 DGMLLAVHKHLQPRKDEPIIEKSVRFRTVGDVTCTGCVESTAATVSEVIA
TH_2243|M.thermoresistible__bu DGMLLAVHRFMQPDPGEEVFETTVRFRTVGDVTCTGCVESTAATVDEVIA
MSMEG_4979|M.smegmatis_MC2_155 DGMLLAVHEFMQPRDGEEVFETSVRFRTVGDVTCTGCVESTAATVEQVIA
MAB_1660c|M.abscessus_ATCC_199 DGMLLTPGPWGGPRDGEELQTLSVRYRTVGDGSTTGAVLSDAADVHAVLA
*****: * .* : :**:***** : **.* * *: * *:*
Mb1316|M.bovis_AF2122/97 ETAVARLTERGATRADDRISEAGMEDRKRQGYF
Rv1285|M.tuberculosis_H37Rv ETAVARLTERGATRADDRISEAGMEDRKRQGYF
MMAR_4133|M.marinum_M ETAVARLTERGATRADDRISESGMEDRKRQGYF
MUL_3996|M.ulcerans_Agy99 ETAVARLTERGATRADDRISESGMEDRKRQGYF
MAV_1436|M.avium_104 ETAVSRLTERGATRADDRISEAGMEDRKRQGYF
Mflv_2271|M.gilvum_PYR-GCK ETAISRLTERGATRADDRISEAGMEDRKREGYF
Mvan_4423|M.vanbaalenii_PYR-1 ETAISRLTERGATRADDRISEAGMEDRKREGYF
TH_2243|M.thermoresistible__bu ETAVSRLTERGATRADDRISEAGMEDRKREGYF
MSMEG_4979|M.smegmatis_MC2_155 ETAVSRLTERGATRADDRISEAGMEDRKREGYF
MAB_1660c|M.abscessus_ATCC_199 EVASSRITERGATRGDDRVSEAAMEDRKREGYF
*.* :*:*******.***:**:.******:***