For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTEFLSQPTLEGPTLTLRPLGEDDLEPLYRAANDPLIWAQHPSSDRYERPVFEKWFAEALAAKSLVIID HSTGEMIGSSRFYEWDPDKREVAIGYTFITREYWGGTVNAELKTLMLDYAFINADLVWFHVAAANLRSQK ALAKIGAHEHHRQKREINGALEDYVYFTITATDWRDASD
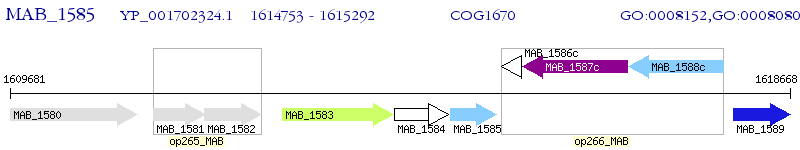
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1585 | - | - | 100% (179) | putative acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2083 | - | 5e-05 | 23.03% (178) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2186 | - | 8e-05 | 28.08% (146) | hypothetical protein MMAR_2186 |
| M. avium 104 | MAV_3396 | - | 2e-07 | 31.94% (144) | acetyltransferase, gnat family protein |
| M. smegmatis MC2 155 | MSMEG_5612 | - | 3e-08 | 27.48% (131) | amino-acid acetyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1771 | - | 5e-05 | 28.08% (146) | hypothetical protein MUL_1771 |
| M. vanbaalenii PYR-1 | Mvan_4944 | - | 0.0001 | 29.25% (106) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5612|M.smegmatis_MC2_155 ---------MKPVTLTGRRWVQLEPLTREHIP-----------------E
Mvan_4944|M.vanbaalenii_PYR-1 -----MSGFVEPVTLTGDRWVSLEPLAREHLP-----------------E
MMAR_2186|M.marinum_M ------MAHFPPRYLDSRRLLLRPPKLDDAGP-----------------I
MUL_1771|M.ulcerans_Agy99 ------MAHFPPRYLDSRRLLLRPPKLDDAGP-----------------I
MAV_3396|M.avium_104 -MNRTAMPEYPPDRITGPRLSLRLPVLDDAGP-----------------L
Mflv_2083|M.gilvum_PYR-GCK MTGHAENEFGQPVGAAVRDWAPVPSPNDDTLAGRYCTLEHLDPDRHADDL
MAB_1585|M.abscessus_ATCC_1997 ----MSTEFLSQPTLEGPTLTLRPLGEDDLEP------------------
: .
MSMEG_5612|M.smegmatis_MC2_155 IDAVAADGELG-TLWYTTTPQPGAAEQWVERMLKLRAADDGLSFVVRDLD
Mvan_4944|M.vanbaalenii_PYR-1 VEAAAADGELG-RLWFTGTPKPGAAGTWVEMRLGLQRPDTGLTFVVRGRD
MMAR_2186|M.marinum_M YKKIAKDPEVTRFLLWRAHRNVAETRRVLINQLSRSRYERNWVISLHG-S
MUL_1771|M.ulcerans_Agy99 YKKIAKDPEVTRFLLWRAHRNVAETRRVLINQLSRSRYERNWVISLHG-S
MAV_3396|M.avium_104 FQRVARDPQVTKYLLWAPHPDVAATRRVITEKLNASADEKTWVIELRH-R
Mflv_2083|M.gilvum_PYR-GCK YAAYAAAPDARDWTYLPVGPFPTAQSYRAWADAAAESRDPRHYAVVDNVT
MAB_1585|M.abscessus_ATCC_1997 LYRAANDP-----LIWAQHPSSDRYERPVFEKWFAEALAAKSLVIIDHST
* :
MSMEG_5612|M.smegmatis_MC2_155 GNLVGSSSYLNVDGTNRRLEIGHTWYTAAARGTGVNAEAKLLLLGHAFDE
Mvan_4944|M.vanbaalenii_PYR-1 GALIGSSSYLNVDGPNRRLEIGNTWFVETARRTGVNTETKLLMLGHAFDE
MMAR_2186|M.marinum_M GEIVGLISCR--SDAPRSIEVG-YCLGRQWWDMGFMSEALSLLLAGLPAD
MUL_1771|M.ulcerans_Agy99 GEIVGLISCR--SDAPRSIEVG-YCLGRQWWDMGFMSEALSLLLAGLPAD
MAV_3396|M.avium_104 GEVVGLLSCR--RTAPHSVEIG-YCLGRRWWGKGLMSEVLGMLLTALDAD
Mflv_2083|M.gilvum_PYR-GCK GRALGTLALMRQDLANGVIEVGFVMFSRALQRTPVSTEAQFLLMRHVFG-
MAB_1585|M.abscessus_ATCC_1997 GEMIGSSRFYEWDPDKREVAIGYTFITREYWGGTVNAELKTLMLD--YAF
* :* : :* . :* :::
MSMEG_5612|M.smegmatis_MC2_155 LNCIAVEFRTHFFNFASRAAIERLGAKQDGVLRSHQIAADGSRRDTVVYS
Mvan_4944|M.vanbaalenii_PYR-1 LGCVAVEFRTHYFNFASRAAIERLGAKLDGVLRSHQLLPDGSRRDTVVYS
MMAR_2186|M.marinum_M AAGCRVWATCHVDNARSARLLQRAGFVLEARLCRHAVYPTMGPEPNDSLL
MUL_1771|M.ulcerans_Agy99 AAGCRVWATCHVDNARSARLLQRAGFVLEARLCRHAVYPTMGPEPNDSLL
MAV_3396|M.avium_104 REVYRVWATCSVDNQRSARLLEHAGFFLEGRLARHAVYPTMGPEPQDSLL
Mflv_2083|M.gilvum_PYR-GCK LGYRRYEWKCDSLNEPSRRTAERLGFTYEGTFRQAVVYK-GRNRDTAWFA
MAB_1585|M.abscessus_ATCC_1997 INADLVWFHVAAANLRSQKALAKIGAHEHHRQKRE---INGALEDYVYFT
* * : * . .
MSMEG_5612|M.smegmatis_MC2_155 ILDIEWPAARSNLNFRLDRRDRNR-------------
Mvan_4944|M.vanbaalenii_PYR-1 ILDIEWPAVRSNLRFRLDRNS----------------
MMAR_2186|M.marinum_M YTRILRLNRAPMPRFAP--------------------
MUL_1771|M.ulcerans_Agy99 YTRILRLNRAPMPRFAP--------------------
MAV_3396|M.avium_104 YAKILR-------------------------------
Mflv_2083|M.gilvum_PYR-GCK MTDGDWSSARAAFEEWLDPANFDGAGRQLGPLRRTRL
MAB_1585|M.abscessus_ATCC_1997 ITATDWRDASD--------------------------