For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGHAENEFGQPVGAAVRDWAPVPSPNDDTLAGRYCTLEHLDPDRHADDLYAAYAAAPDARDWTYLPVGP FPTAQSYRAWADAAAESRDPRHYAVVDNVTGRALGTLALMRQDLANGVIEVGFVMFSRALQRTPVSTEAQ FLLMRHVFGLGYRRYEWKCDSLNEPSRRTAERLGFTYEGTFRQAVVYKGRNRDTAWFAMTDGDWSSARAA FEEWLDPANFDGAGRQLGPLRRTRL
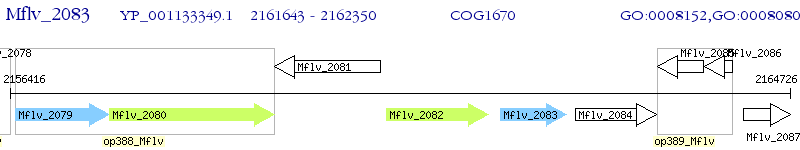
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2083 | - | - | 100% (235) | GCN5-related N-acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4621c | - | 3e-05 | 32.62% (141) | putative acetyltransferase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5612 | - | 3e-10 | 30.65% (124) | amino-acid acetyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4944 | - | 4e-10 | 28.95% (190) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5612|M.smegmatis_MC2_155 ---------MKPVTLTGRRWVQLEPLTREHIP----EIDAVAADGELGTL
Mvan_4944|M.vanbaalenii_PYR-1 -----MSGFVEPVTLTGDRWVSLEPLAREHLP----EVEAAAADGELGRL
Mflv_2083|M.gilvum_PYR-GCK MTGHAENEFGQPVGAAVRDWAPVPSPNDDTLAGRYCTLEHLDPDRHADDL
MAB_4621c|M.abscessus_ATCC_199 ---------MEPVELQTDRLMLRAVADADAAA----IVDACNDPEILHYL
:** : . :: *
MSMEG_5612|M.smegmatis_MC2_155 WYTTTPQPGAAEQWVERMLK---------------LRAADDGLSFVVRDL
Mvan_4944|M.vanbaalenii_PYR-1 WFTGTPKPGAAGTWVEMRLG---------------LQRPDTGLTFVVRGR
Mflv_2083|M.gilvum_PYR-GCK YAAYAAAP-DARDWTYLPVGPFPTAQSYRAWADAAAESRDPRHYAVVDNV
MAB_4621c|M.abscessus_ATCC_199 PLPVPYTVGDARSFIAASAQ-------------EWVAGTRYGFGFFVMD-
. . * : .* .
MSMEG_5612|M.smegmatis_MC2_155 DGNLVGSSSYLNVDGTNRRLEIGHTWYTAAARGTGVNAEAKLLLLGHAFD
Mvan_4944|M.vanbaalenii_PYR-1 DGALIGSSSYLNVDGPNRRLEIGNTWFVETARRTGVNTETKLLMLGHAFD
Mflv_2083|M.gilvum_PYR-GCK TGRALGTLALMRQDLANGVIEVGFVMFSRALQRTPVSTEAQFLLMRHVFG
MAB_4621c|M.abscessus_ATCC_199 TGELAGTCSLKPVT--SGVLEVG-YWSAAQHRRKGLTTEAVQRLCQWGFD
* *: : . :*:* : . :.:*: : *.
MSMEG_5612|M.smegmatis_MC2_155 ELNCIAVEFRTHFFNFASRAAIERLGAKQDGVLRSHQIAADGSRRDTVVY
Mvan_4944|M.vanbaalenii_PYR-1 ELGCVAVEFRTHYFNFASRAAIERLGAKLDGVLRSHQLLPDGSRRDTVVY
Mflv_2083|M.gilvum_PYR-GCK -LGYRRYEWKCDSLNEPSRRTAERLGFTYEGTFR-QAVVYKGRNRDTAWF
MAB_4621c|M.abscessus_ATCC_199 VLDAHRIEWWAVVGNEGSRAVAEAAGFEMEGLLRQRACRRQEP-------
*. *: * ** . * * :* :* : .
MSMEG_5612|M.smegmatis_MC2_155 SILDIEWPAARSNLNFRLDRRDRNR-------------
Mvan_4944|M.vanbaalenii_PYR-1 SILDIEWPAVRSNLRFRLDRNS----------------
Mflv_2083|M.gilvum_PYR-GCK AMTDGDWSSARAAFEEWLDPANFDGAGRQLGPLRRTRL
MAB_4621c|M.abscessus_ATCC_199 ----ADWWVGG-----LLPRPE----------------
:* * .