For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AHFPPRYLDSRRLLLRPPKLDDAGPIYKKIAKDPEVTRFLLWRAHRNVAETRRVLINQLSRSRYERNWVI SLHGSGEIVGLISCRSDAPRSIEVGYCLGRQWWDMGFMSEALSLLLAGLPADAAGCRVWATCHVDNARSA RLLQRAGFVLEARLCRHAVYPTMGPEPNDSLLYTRILRLNRAPMPRFAP*
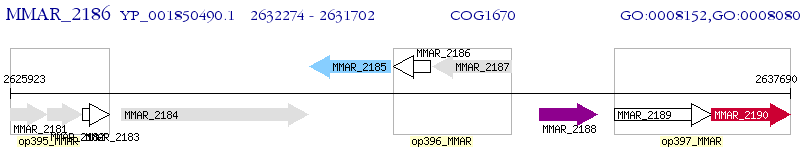
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2186 | - | - | 100% (190) | hypothetical protein MMAR_2186 |
| M. marinum M | MMAR_0919 | rimL | 1e-29 | 36.87% (179) | acetyltransferase, RimL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1585 | - | 7e-05 | 28.08% (146) | putative acetyltransferase |
| M. avium 104 | MAV_3396 | - | 3e-62 | 60.34% (179) | acetyltransferase, gnat family protein |
| M. smegmatis MC2 155 | MSMEG_4972 | - | 7e-39 | 45.81% (179) | acetyltransferase |
| M. thermoresistible (build 8) | TH_2247 | - | 2e-41 | 46.07% (178) | acetyltransferase |
| M. ulcerans Agy99 | MUL_1771 | - | 1e-111 | 100.00% (190) | hypothetical protein MUL_1771 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2186|M.marinum_M -----MAHFPPRYLDSRRLLLRPPKLDDAGPIYKKIAKDPEVTRFLLWRA
MUL_1771|M.ulcerans_Agy99 -----MAHFPPRYLDSRRLLLRPPKLDDAGPIYKKIAKDPEVTRFLLWRA
MAV_3396|M.avium_104 MNRTAMPEYPPDRITGPRLSLRLPVLDDAGPLFQRVARDPQVTKYLLWAP
MSMEG_4972|M.smegmatis_MC2_155 --MGTMLHVPT--LTGPRVRLREPVPDDAEALYTRVASDPEVTRYLSWKP
TH_2247|M.thermoresistible__bu VQDEVMTTVET--LAGPRVRLRPPRTDDADTLFSRVCSDPQVARFVSWRP
MAB_1585|M.abscessus_ATCC_1997 ---MSTEFLSQPTLEGPTLTLRPLGEDDLEPLYR-AANDP-----LIWAQ
: . : ** ** .:: . ** : *
MMAR_2186|M.marinum_M HRNVAETRRVLINQLSRSR-----------YERNWVISLHGSGEIVGLIS
MUL_1771|M.ulcerans_Agy99 HRNVAETRRVLINQLSRSR-----------YERNWVISLHGSGEIVGLIS
MAV_3396|M.avium_104 HPDVAATRRVITEKLNASA-----------DEKTWVIELRHRGEVVGLLS
MSMEG_4972|M.smegmatis_MC2_155 HADVAETRRVITTLFNIGS------------EESWLIEADG--EVVGLCG
TH_2247|M.thermoresistible__bu HVDVRETRRVIDTIFNANARAGSKADSGADTDRTWVMDLTGV-GVVGLCG
MAB_1585|M.abscessus_ATCC_1997 HPSSDRYERPVFEKWFAEA----------LAAKSLVIIDHSTGEMIGSSR
* . .* : .. :: ::*
MMAR_2186|M.marinum_M CRSDAP--RSIEVGYCLG-RQWWDMGFMSEALSLLLAGLPADAAGCRVWA
MUL_1771|M.ulcerans_Agy99 CRSDAP--RSIEVGYCLG-RQWWDMGFMSEALSLLLAGLPADAAGCRVWA
MAV_3396|M.avium_104 CRRTAP--HSVEIGYCLG-RRWWGKGLMSEVLGMLLTALDADREVYRVWA
MSMEG_4972|M.smegmatis_MC2_155 RRRPQA--HEVELGYCLG-RPWWRRGLMSEAMSVLIDDTERDPRVYRISA
TH_2247|M.thermoresistible__bu WRRPEP--HAVEFGYCLG-RRWWGRGLASEALSLLIDAVSRDERVYRLWA
MAB_1585|M.abscessus_ATCC_1997 FYEWDPDKREVAIGYTFITREYWGGTVNAELKTLMLDYAFIN--ADLVWF
. : : .** : * :* . :* ::: : :
MMAR_2186|M.marinum_M TCHVDNARSARLLQRAGFVLEARLCRHAVYPTMGPEPNDSLLYTRILRLN
MUL_1771|M.ulcerans_Agy99 TCHVDNARSARLLQRAGFVLEARLCRHAVYPTMGPEPNDSLLYTRILRLN
MAV_3396|M.avium_104 TCSVDNQRSARLLEHAGFFLEGRLARHAVYPTMGPEPQDSLLYAKILR--
MSMEG_4972|M.smegmatis_MC2_155 YCHVDNAGSAGVLRHCGLTLEGRLVRYSVFPNLGNEPQDVLMFGKAVR--
TH_2247|M.thermoresistible__bu VCHVDNAASAAVLRRCGLTLEGRLARYAVLPNLGSEPQDVLLFAKAVR--
MAB_1585|M.abscessus_ATCC_1997 HVAAANLRSQKALAKIGAHEHHRQKR-----EINGALEDYVYFTITATDW
. * * * : * . * * :. :* : :
MMAR_2186|M.marinum_M RAPMPRFAP
MUL_1771|M.ulcerans_Agy99 RAPMPRFAP
MAV_3396|M.avium_104 ---------
MSMEG_4972|M.smegmatis_MC2_155 ---------
TH_2247|M.thermoresistible__bu ---------
MAB_1585|M.abscessus_ATCC_1997 RDASD----