For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAPKRAPEYGPAGSARSSLSAVNTLVRGLPLSDGAAVEVIEDAASLAGRLLGFGVRMTVRTVLQIGSHA PTFPYPFGLIEELGRVLVPPRGTVKATVTLPNTNARLIRAKGVGPADGTGRVVLYLHGGAFIVGGPNTHS AMVSTISQHADAPCLVVNYRMPPKASLDQAIDDCLDGYRWLRAQGYAAEQIVLAGDSAGGFLSVAVAERL LAEDGEAPAALALISPLIELDPAAKVAHENARTDVMLPPNCFEALEKILTKAGGGIPPQEIIDKVDGRMP QTLVHCSGSEVLLYDARLLGKRLAEQGVPVEIKIWPGQMHVFQVATKVVPEAKRSLAQIGQYIRDAVPGT FPAEFAAPAAV
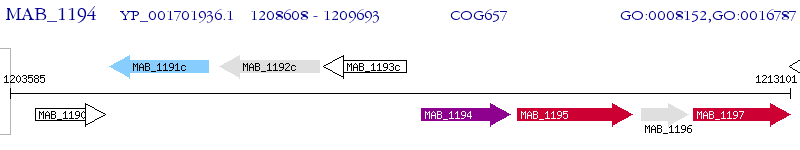
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1194 | - | - | 100% (361) | putative lipase LipU |
| M. abscessus ATCC 19977 | MAB_4044c | - | 1e-50 | 36.31% (325) | putative hydrolase/esterase/lipase |
| M. abscessus ATCC 19977 | MAB_0141c | - | 5e-36 | 32.31% (294) | putative esterase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1105 | lipU | 4e-86 | 53.38% (296) | putative lipase LIPU |
| M. gilvum PYR-GCK | Mflv_2042 | - | 1e-98 | 51.68% (358) | alpha/beta hydrolase domain-containing protein |
| M. tuberculosis H37Rv | Rv1076 | lipU | 4e-86 | 53.38% (296) | lipase LipU |
| M. leprae Br4923 | MLBr_00314 | - | 2e-94 | 52.78% (324) | putative esterase |
| M. marinum M | MMAR_4391 | lipU | 6e-97 | 51.69% (354) | lipase LipU |
| M. avium 104 | MAV_1200 | - | 1e-94 | 50.14% (353) | alpha/beta hydrolase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5271 | - | 1e-99 | 52.97% (353) | esterase |
| M. thermoresistible (build 8) | TH_1370 | lipF | 2e-99 | 52.12% (353) | PROBABLE ESTERASE/LIPASE LIPF |
| M. ulcerans Agy99 | MUL_0204 | lipU | 6e-86 | 53.95% (291) | lipase LipU |
| M. vanbaalenii PYR-1 | Mvan_4675 | - | 8e-99 | 51.68% (358) | alpha/beta hydrolase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1105|M.bovis_AF2122/97 --------------------------------------------------
Rv1076|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4391|M.marinum_M ---MTAPSRVPSNPIHAIRPRDVLKARRLEARRFAISDGAPVEVVESGPS
MUL_0204|M.ulcerans_Agy99 --------------------------------------------------
MAV_1200|M.avium_104 ---MTAPSEVSGSQS-PVRVHDVLKARRARIRKFAISDGAPVEVLESGPS
MLBr_00314|M.leprae_Br4923 ---------------------------------MAISAGAPVEIVESGPS
Mflv_2042|M.gilvum_PYR-GCK ---MTAPSKVPATPHVAINA-----ARGRKQRRFPVSDGAPVEVVEDGPS
Mvan_4675|M.vanbaalenii_PYR-1 ---MTAPSKVPATPHAAIAA-----ARAGKIRRFPVSDGAPVEVVEDGPS
TH_1370|M.thermoresistible__bu ---MTAPSKVPRTSPLATRA-----ARGHYSRTFPVSDGTPSEYVEDGPS
MSMEG_5271|M.smegmatis_MC2_155 MVAMTAPSKVTRSPRAGIGP-----GGAYRTRKFPVSDSAPVEIVEDGTS
MAB_1194|M.abscessus_ATCC_1997 ---MTAPKRAPEYGPAGSAR-SSLSAVNTLVRGLPLSDGAAVEVIEDAAS
Mb1105|M.bovis_AF2122/97 -----------MAVRPVLAVGSYLPHAPWPWGVIDQAARVLLPASTTVRA
Rv1076|M.tuberculosis_H37Rv -----------MAVRPVLAVGSYLPHAPWPWGVIDQAARVLLPASTTVRA
MMAR_4391|M.marinum_M VAARVATLASRLTIRPVLAVGSHVPHLPWPWGLIDHAAKALVPASATVRA
MUL_0204|M.ulcerans_Agy99 ----------------MLAVGSHVPHLPWPWGLIDHAAKALVPASATVRA
MAV_1200|M.avium_104 VAARLANLTSRLTIRPILAVGSHAPNLPWPWGLIDLTARVLLPVSATVRE
MLBr_00314|M.leprae_Br4923 IAARLASLTSRLTIRPILAVGSYVPQLPWPFGLIDHAARVLLPVTNAARA
Mflv_2042|M.gilvum_PYR-GCK VAGRVLGLAAMATIKPFLTVGSYAPKLPWPWGLVDFAARVLKPAPGTVRA
Mvan_4675|M.vanbaalenii_PYR-1 IAGRLMGLAAVATVKPFLTVGSYAPKLPWPWGLLDFAARVLKPAPGTVRA
TH_1370|M.thermoresistible__bu LAGRLTALAAYLTVRPTLAIGSFAPRLPWPWGLLDFAARAVRPVPGTVRA
MSMEG_5271|M.smegmatis_MC2_155 VAGRLASLAAMLTIRPTLAIGSRVPHLPWPFGALNFAARMLRPSPGTIKA
MAB_1194|M.abscessus_ATCC_1997 LAGRLLGFGVRMTVRTVLQIGSHAPTFPYPFGLIEELGRVLVPPRGTVKA
* :** * *:*:* :: .: : * : :
Mb1105|M.bovis_AF2122/97 AVSLPNASAQLVRASGVLPADGTRRAVLYLHGGAFLTCGANSHGRLVELL
Rv1076|M.tuberculosis_H37Rv AVSLPNASAQLVRASGVLPADGTRRAVLYLHGGAFLTCGANSHGRLVELL
MMAR_4391|M.marinum_M AVSLPNAAAQLVRAPGVLPADGTRRVVLYLHGGAFLTCGANSHGRLVELL
MUL_0204|M.ulcerans_Agy99 AVSLPNAAAQLVRAPGVLPADGTRRVVLYLHGGAFLTCGANSHGRFFELL
MAV_1200|M.avium_104 TVALPHASAQLVRAPGVLPADGTRRVVVYLHGGAFLTCGANSHGRLVEAL
MLBr_00314|M.leprae_Br4923 EVNLSNTSAQLVRAAGVRPADGSGRIVLYLHGGAFLGCGANSHRRLVETL
Mflv_2042|M.gilvum_PYR-GCK TIKLPHCTAQLVRAAGVLPADGNRTVILYLHGGAFLTCGVNTHGRLVTAL
Mvan_4675|M.vanbaalenii_PYR-1 TIGLPHCNAQLVRAAGVLPADGHRSVILYLHGGAFLTCGVNTHGRLVTAL
TH_1370|M.thermoresistible__bu TIRLPHCTAQLVRAPGVLPADGRRSVVLYLHGGAFLTCGAHTHGRLVTAL
MSMEG_5271|M.smegmatis_MC2_155 TIALPNCTAQLIRADGVLPADGTRSVILYLHGGAFLACGSNTHGGMATSL
MAB_1194|M.abscessus_ATCC_1997 TVTLPNTNARLIRAKGVGPADGTGRVVLYLHGGAFIVGGPNTHSAMVSTI
: *.: *:*:** ** **** ::*******: * ::* : :
Mb1105|M.bovis_AF2122/97 SKFADSPVLVVDYRLIPKHSIGMALDDCHDGYRWLRLLGYEPEQIVLAGD
Rv1076|M.tuberculosis_H37Rv SKFADSPVLVVDYRLIPKHSIGMALDDCHDGYRWLRLLGYEPEQIVLAGD
MMAR_4391|M.marinum_M SKFADAPVLIVNYRLMPKHSIGMALDDCHDGYRWLRLLGYDPEQIVLAGD
MUL_0204|M.ulcerans_Agy99 SKFADAPVLIVNYRLMPKHSIGMALDDCHDGYRWLRLLGYDPEQIVLAGD
MAV_1200|M.avium_104 STFADAPVLVVNYRLLPKNSVGMALQDCHDAYQWLRLRGYQPDQIVLAGD
MLBr_00314|M.leprae_Br4923 SKLADSPILVVNYRLLPKHSIGMALDDCHDGYQWLRRLGYDPEQIVLAGD
Mflv_2042|M.gilvum_PYR-GCK SKCADAPALVVNYRMIPKHSVGTALDDCYDAYKWLRETGYEPEQIVLAGD
Mvan_4675|M.vanbaalenii_PYR-1 SKCADAPVLVVNYRMIPKHSVGTALDDCFDGYKWLRELGYEPDQIVLAGD
TH_1370|M.thermoresistible__bu SEFADSPVLVVNYRMIPKHSVGTAIDDCYDGYQWLRAKGYEPDQIVLAGD
MSMEG_5271|M.smegmatis_MC2_155 SEYADSPVLVVDYRMVPKHSVGTAVDDCYDAYVWLRRAGYEPEQIVVAGD
MAB_1194|M.abscessus_ATCC_1997 SQHADAPCLVVNYRMPPKASLDQAIDDCLDGYRWLRAQGYAAEQIVLAGD
* **:* *:*:**: ** *:. *::** *.* *** ** .:***:***
Mb1105|M.bovis_AF2122/97 SAGGYLALALAQRLQEV------GEEPAALVAISPLLQLAKEHKQAHPNI
Rv1076|M.tuberculosis_H37Rv SAGGYLALALAQRLQEV------GEEPAALVAISPLLQLAKEHKQAHPNI
MMAR_4391|M.marinum_M SAGGYLALTLAQRLQEE------GEEPAALVAISPLLQLAKEQKKAHPNI
MUL_0204|M.ulcerans_Agy99 SAGGYLALTLAQRLQEE------GEEPAALVAISPLLQLAKEQKKAHPNI
MAV_1200|M.avium_104 SAGGYLALTLAQRLQRD------GEEPAALVMISPLLQLAKEPKQAHPNI
MLBr_00314|M.leprae_Br4923 SAGGYLALALAQRLQDV------GEEPAALVAISPLLQLAKKGKQAHPNA
Mflv_2042|M.gilvum_PYR-GCK SAGGYLALALAEKLQAEGINGYVGETPAAVVTMSPLFEIDNEARADHPNV
Mvan_4675|M.vanbaalenii_PYR-1 SAGGYLALSLAERLQAEGINGFVGETPAAIVTMSPLFEIDNEARADHPNV
TH_1370|M.thermoresistible__bu SAGGYLALSLAQRLLDE------GEEPAALVALSPLFEIDNESRAQHPNI
MSMEG_5271|M.smegmatis_MC2_155 SAGGYLGLALAERLLDD------GEVPAALVAMSPLYEIDNRARAEHPNA
MAB_1194|M.abscessus_ATCC_1997 SAGGFLSVAVAERLLAED-----GEAPAALALISPLIELDPAAKVAHENA
****:*.:::*::* ** ***:. :*** :: : * *
Mb1105|M.bovis_AF2122/97 KTDAMFPARAFDALDALVASAAARNQVDGEPEELYEPLEHITPGLPRTLI
Rv1076|M.tuberculosis_H37Rv KTDAMFPARAFDALDALVASAAARNQVDGEPEELYEPLEHITPGLPRTLI
MMAR_4391|M.marinum_M KRDAMFPPKAFDALGELVASAAAKNKIDGKPEELYEPLEHIRPGLPRTLI
MUL_0204|M.ulcerans_Agy99 KRDAMFPPKAFDALGELVASAAAKNRVDGKPEELYEPLEHIRPGLPRTLI
MAV_1200|M.avium_104 DTDAMFGAGAFDALAELVAGAAAKNIVDGKPEEIYEPLEHIEPGLPRTLI
MLBr_00314|M.leprae_Br4923 KTDAMFPPKAFDALGRLVASSAAKNKVDDKPEELYEPLDHIAPGLPRTLI
Mflv_2042|M.gilvum_PYR-GCK RTDSMFPPKAFHALVELIEEAAARRMVNGAPEEVYEPLDHIEPGLPRTLI
Mvan_4675|M.vanbaalenii_PYR-1 RTDAMFPPKAFHALVELIEAAAERRMVDGKPEEVYEPLDHIEPGLPRTLI
TH_1370|M.thermoresistible__bu RTDAMFPPRAFSALIELVEEAAERRTTNGRPEEVYEPLDNIEPGLPRTLI
MSMEG_5271|M.smegmatis_MC2_155 RRDAMFPPKAFDALVEIVEAAAAR---NG--EQVYEPLDHIEPGLPRTLI
MAB_1194|M.abscessus_ATCC_1997 RTDVMLPPNCFEALEKILTKAGGG-------IPPQEIIDKVDGRMPQTLV
* *: . .* ** :: :. * :::: :*:**:
Mb1105|M.bovis_AF2122/97 HVSGSEVLLHDAQLAAAKLAAAGVPAEVRVWPGQVHDFQVAASMLPEAIR
Rv1076|M.tuberculosis_H37Rv HVSGSEVLLHDAQLAAAKLAAAGVPAEVRVWPGQVHDFQVAASMLPEAIR
MMAR_4391|M.marinum_M HVSGSEVLLHDAQLAARKLAAVGVPAEVRVWPGQVHDFQVAGPLLPEATR
MUL_0204|M.ulcerans_Agy99 HVSGSEVLLHDAQLAARKLAAVGVPAEVRVWPGQVHDFQVAGPLLPEATR
MAV_1200|M.avium_104 HVSGSEVLLHDARLAASRLAAAGVPAEVRVWPGQIHDFQLAAPMVPEARR
MLBr_00314|M.leprae_Br4923 HVSGSEVLLHDAQLAATKLAAVGVPAEIRVWPGQVHVFQAAAPMVPEATL
Mflv_2042|M.gilvum_PYR-GCK HVSGSEVLLSDARKAARLLAASGVPVEVRIWPGQMHVFQLGAPMVPEASR
Mvan_4675|M.vanbaalenii_PYR-1 HVSGSEVLLSDARKAARLLAAAGVPVEVRVWPGQMHVFQLGAPAVSEANR
TH_1370|M.thermoresistible__bu HVSGSEALISDARKAARRLAAAGVPVEIRVWPGQMHVFQIAAPFVPEAKR
MSMEG_5271|M.smegmatis_MC2_155 HVSGSEVLISDARKAARMLCAAGVPVEVHVWPGQMHVFQIAGNLVPEAKR
MAB_1194|M.abscessus_ATCC_1997 HCSGSEVLLYDARLLGKRLAEQGVPVEIKIWPGQMHVFQVATKVVPEAKR
* ****.*: **: . *. ***.*:::****:* ** . :.**
Mb1105|M.bovis_AF2122/97 SLRQIGEYIREATG-------------
Rv1076|M.tuberculosis_H37Rv SLRQIGEYIREATG-------------
MMAR_4391|M.marinum_M SLRQIGDYIREATG-------------
MUL_0204|M.ulcerans_Agy99 SLRQIGDYIREATG-------------
MAV_1200|M.avium_104 SLRQIGDYIREATG-------------
MLBr_00314|M.leprae_Br4923 SLRQISQYIREATASKASRDRQSL---
Mflv_2042|M.gilvum_PYR-GCK SIKQIGEYIREATW-------------
Mvan_4675|M.vanbaalenii_PYR-1 SIKQIGEYIREATW-------------
TH_1370|M.thermoresistible__bu SLRQIGEYIREATW-------------
MSMEG_5271|M.smegmatis_MC2_155 SLRQIGAYIREATW-------------
MAB_1194|M.abscessus_ATCC_1997 SLAQIGQYIRDAVPGTFPAEFAAPAAV
*: **. ***:*.