For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLVLVLIGILAGLTTVLFGFGGGFVTVPVIVWADGALGAAAPAVAVATSAVVMVVNAAAATAATSRATLA HLRSAVPLVALLGCGGALGAGLARLAPPQLTTWGFVGYLVITIADVLLRPGFLRPPAVIDGPRREFAIPA GLGIPIGATASFLGVGGSVMTVPLLRRGGQPMSVATALANPLTLAISVPALAVFLTSTAAVAGEPYFAGA VDCRAAAALLAGALPLVMWLRRRPPRLPDTVHAWAYVGLLVVSAVAMLTAGAP
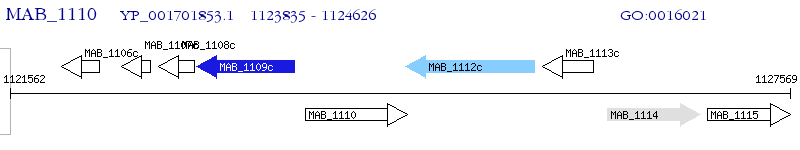
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1110 | - | - | 100% (263) | hypothetical protein MAB_1110 |
| M. abscessus ATCC 19977 | MAB_1170 | - | 1e-09 | 28.81% (236) | hypothetical protein MAB_1170 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1031c | - | 6e-06 | 28.19% (227) | hypothetical protein Mb1031c |
| M. gilvum PYR-GCK | Mflv_0983 | - | 3e-10 | 28.12% (224) | hypothetical protein Mflv_0983 |
| M. tuberculosis H37Rv | Rv1004c | - | 7e-06 | 28.19% (227) | hypothetical protein Rv1004c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1775 | - | 5e-05 | 32.72% (162) | hypothetical protein MAV_1775 |
| M. smegmatis MC2 155 | MSMEG_5387 | - | 9e-10 | 24.56% (228) | hypothetical protein MSMEG_5387 |
| M. thermoresistible (build 8) | TH_4293 | - | 1e-10 | 26.14% (264) | PUTATIVE putative protein of unknown function DUF81 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1828 | - | 7e-11 | 27.59% (203) | hypothetical protein Mvan_1828 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0983|M.gilvum_PYR-GCK -------MMALTVGLAVFVGIALGLLGG--GGSILTVPLLAYVAG-----
Mvan_1828|M.vanbaalenii_PYR-1 -------MIALTIGLAVFVGIALGLLGG--GGSILTVPLLAYVAG-----
TH_4293|M.thermoresistible__bu -------MIGLIIALAVLVGVALGLLGG--GGSILMVPLLAYVAG-----
MSMEG_5387|M.smegmatis_MC2_155 -------MIALTIVLAVFVGIALGMLGG--GGSILTVPLLAYVAG-----
Mb1031c|M.bovis_AF2122/97 MSISCRVREGFVMRLAIVGTAAAAAIGGTLAVAPLTLSTPERVAGGTCSA
Rv1004c|M.tuberculosis_H37Rv MSISCRVREGFVMRLAIVGTAAAAAIGGTLAVAPLTLSTPERVAGGTCSA
MAB_1110|M.abscessus_ATCC_1997 --------MLVLVLIGILAGLTTVLFGF--GGGFVTVPVIVWADG-----
MAV_1775|M.avium_104 ------MAAYVRAQPDAVRVIAVATFAS-------SVPLALYAAT-----
. . : :. :. .
Mflv_0983|M.gilvum_PYR-GCK ---------------------MDAKQAIATSLLVVGVTSAIGAVS--HAR
Mvan_1828|M.vanbaalenii_PYR-1 ---------------------MDAKAAIATSLLVVGVTSAIGAIS--HAR
TH_4293|M.thermoresistible__bu ---------------------MDAKAAIATSLLVVGVTSAVGAVS--HAR
MSMEG_5387|M.smegmatis_MC2_155 ---------------------MDAKQAVTTSLLVVGVTSAVSAVS--HAR
Mb1031c|M.bovis_AF2122/97 GQQCDRLAAVLMPDTATPSGPAAAEHAVPAPFEPVADTIAPGLVPRPGVP
Rv1004c|M.tuberculosis_H37Rv GQQCDRLAAVLMPDTATPSGPAAAEHAVPAPFEPVADTIAPGLVPRPGVP
MAB_1110|M.abscessus_ATCC_1997 -------------------ALGAAAPAVAVATSAVVMVVNAAAATAATSR
MAV_1775|M.avium_104 ---------------------------AAARLRRLGAGPTTAAIT-----
.. : . .
Mflv_0983|M.gilvum_PYR-GCK AGRVQWRTG--------LIFGAAG--------------------------
Mvan_1828|M.vanbaalenii_PYR-1 AGRVQWRTG--------LIFGAAG--------------------------
TH_4293|M.thermoresistible__bu AGQVRWRTG--------LIFGAAG--------------------------
MSMEG_5387|M.smegmatis_MC2_155 AGRVQWRAG--------LSFGAAG--------------------------
Mb1031c|M.bovis_AF2122/97 AAAAVPRVGPPAVPGLPNIPGAAGPALPPPPALPNLAAPSVPGVGIPGIG
Rv1004c|M.tuberculosis_H37Rv AAAAVPRVGPPAVPGLPNIPGAAGPALPPPPALPNLAAPSVPGVGIPGIG
MAB_1110|M.abscessus_ATCC_1997 ATLAHLRSAVP-------LVALLG--------------------------
MAV_1775|M.avium_104 LTGGTLAAG---------LLALAG--------------------------
. . *
Mflv_0983|M.gilvum_PYR-GCK ----------------------MAGAYAGGLLAR--FIPGTVLLIGFALM
Mvan_1828|M.vanbaalenii_PYR-1 ----------------------MAGAYAGGLLAR--YIPGTILLIGFAVM
TH_4293|M.thermoresistible__bu ----------------------MAGAYAGGILAR--FIPGTVLLLGFAVM
MSMEG_5387|M.smegmatis_MC2_155 ----------------------MAGAYAGGVLGR--FIPGTALLIAFAVV
Mb1031c|M.bovis_AF2122/97 IPGIGIPGIGIPGVPDPITGVNTAAAVVNGVLGVGGTAAGVVTASAVAVT
Rv1004c|M.tuberculosis_H37Rv IPGIGIPGIGIPGVPDPITGVNTAAAVVNGVLGVGGTAAGVVTASAVAVT
MAB_1110|M.abscessus_ATCC_1997 -----------------------CGGALGAGLAR--LAPPQLTTWGFVGY
MAV_1775|M.avium_104 --------------------------LLGWTLSR-----PAVTADTAVVA
. *. .
Mflv_0983|M.gilvum_PYR-GCK MIATAVAMLRGRKNVQATEGS------HRLPVPKILAEGLLVGLVTGLVG
Mvan_1828|M.vanbaalenii_PYR-1 MIATAIAMLRGRKTIETGETG------HRLPVPKIVAEGLIVGLVTGLVG
TH_4293|M.thermoresistible__bu MAATAVAMLRGRRNTATPDTGG----DHRLPVPRILADGLVVGLVTGLVG
MSMEG_5387|M.smegmatis_MC2_155 MVAAAIPMLRGRR---APEATT----AHGLPVVRVVSLGLAVGGLTGMIG
Mb1031c|M.bovis_AF2122/97 YLVLAVNALESSGILPTARGTASTVASLLLPGAQSAAAALPAVGLPALPG
Rv1004c|M.tuberculosis_H37Rv YLVLAVNALESSGILPTARGTASTVASLLLPGAQSAAAALPAVGLPALPG
MAB_1110|M.abscessus_ATCC_1997 LVITIADVLLRPGFLRPPAVID----GPRREFAIPAGLGIPIGATASFLG
MAV_1775|M.avium_104 ALYLLVFLTGGPG--------------------HIVALGVLVAAMA-VPG
. .: . . *
Mflv_0983|M.gilvum_PYR-GCK AG-GGFLVVPALALLGG-LPMPVAVGTS-LIVIAMKSFAGLGGYLSSVQI
Mvan_1828|M.vanbaalenii_PYR-1 AG-GGFLVVPALALLGG-LPMPIAVGTS-LIVIAMKSFAGLAGYLTSVHI
TH_4293|M.thermoresistible__bu AG-GGFLVVPALALLGG-LSMPAAVGTS-LVVIAMKSFAGLAGYLTSVQI
MSMEG_5387|M.smegmatis_MC2_155 AG-GGFLVVPALTLLGG-LPMPAAVGTS-LIVIALNSFSGLAGHLTGLQI
Mb1031c|M.bovis_AF2122/97 VTPASLLAMAAAAGLPG-VGFPSLPGVSPTDLMAMAAAAGLPTSLPGLAG
Rv1004c|M.tuberculosis_H37Rv VTPASLLAMAAAAGLPG-VGFPSLPGVSPTDLMAMAAAAGLPTSLPGLAG
MAB_1110|M.abscessus_ATCC_1997 VG-GSVMTVPLLRRGGQPMSVATALANPLTLAISVPALAVFLTSTAAVAG
MAV_1775|M.avium_104 LTRGVLPRPVARAGLAT-----AALAGSATAVLIWPALGVILPVARVAAL
. . . . : : . :
Mflv_0983|M.gilvum_PYR-GCK DWSLALAVTAAAVVGALLGARLTS-MVNPDSLRKAFGWFVLAMSSVILAQ
Mvan_1828|M.vanbaalenii_PYR-1 NWTLALAVTAAAVIGALIGARLTA-MINPDSLRKTFGWFVLVMSSVILAQ
TH_4293|M.thermoresistible__bu DWPVALAVTVAAVAGALLGSRLTA-LVNPDTLRQAFGWFVLTMSAVILAQ
MSMEG_5387|M.smegmatis_MC2_155 DWALAAAVTGAAVAGCLIGTRLIT-KINADALRKAFGWFVLAMSSVILAQ
Mb1031c|M.bovis_AF2122/97 MSPAELTALVAGGLPMLAAAGLPAGLAGVDPATLAAALPALAAGGLPPGL
Rv1004c|M.tuberculosis_H37Rv MSPAELTALVAGGLPMLAAAGLPAGLAGVDPATLAAALPALAAGGLPPGL
MAB_1110|M.abscessus_ATCC_1997 EPYFAGAVDCRAAAALLAGALPLVMWLRRRPPRLPDTVHAWAYVGLLVVS
MAV_1775|M.avium_104 AWLLAAGIALRDKG------------YGDRGGRAQEGAST----------
.
Mflv_0983|M.gilvum_PYR-GCK EIHLAVGIAGAALTAIAALMTFACSRYAHCPLRRITGIGAARGAHA----
Mvan_1828|M.vanbaalenii_PYR-1 EIHPAVGITAAALTLIAAATTLACQRYAHCPLSRLTTRLTTPKAAA----
TH_4293|M.thermoresistible__bu EAHPAVGAAAAVLTAVAAALTFTCSRYGRCPLRRVAEALSSRPATA----
MSMEG_5387|M.smegmatis_MC2_155 EIHPFVGLAAASLTVIAASMTMVCARFSRCPLRRVTRLSMARPAVA----
Mb1031c|M.bovis_AF2122/97 PALPGVDPAALAAALPALAAGLPALPAGLPPLPAVPALPAPPPLPGPPPL
Rv1004c|M.tuberculosis_H37Rv PALPGVDPAALAAALPALAAGLPALPAGLPPLPAVPALPAPPPLPGPPPL
MAB_1110|M.abscessus_ATCC_1997 AVAMLTAGAP----------------------------------------
MAV_1775|M.avium_104 --------------------------------------------------
Mflv_0983|M.gilvum_PYR-GCK --------------------
Mvan_1828|M.vanbaalenii_PYR-1 --------------------
TH_4293|M.thermoresistible__bu --------------------
MSMEG_5387|M.smegmatis_MC2_155 --------------------
Mb1031c|M.bovis_AF2122/97 PALPSRLCTPGFGPIGVCIP
Rv1004c|M.tuberculosis_H37Rv PALPSRLCTPGFGPIGVCIP
MAB_1110|M.abscessus_ATCC_1997 --------------------
MAV_1775|M.avium_104 --------------------