For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VCAALGVGLLSGGAAVAHADPAFPDLSGFAVVPPDGFFVTNENSSVRSIHFSTPDGIGCMFRASPNMTPT SRQRLSCDGAVPGIPGDAPNAPNVPGIGGASANVPGLGGVNIPGVGTCPMGTVAQKGDGAFEISKGAWSC GTKPEAPLLNVGQKLTYGNVTCAVGAGSVTACQVVTGDQKHGFVLQPSGSTSF
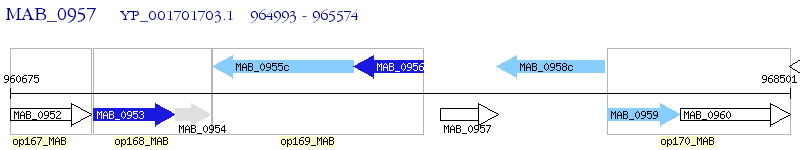
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0957 | - | - | 100% (193) | hypothetical protein MAB_0957 |
| M. abscessus ATCC 19977 | MAB_3472 | - | 8e-14 | 32.51% (203) | hypothetical protein MAB_3472 |
| M. abscessus ATCC 19977 | MAB_3917c | - | 8e-06 | 25.13% (187) | hypothetical protein MAB_3917c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3413 | - | 8e-07 | 24.04% (208) | hypothetical protein Mflv_3413 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3690 | - | 3e-23 | 36.84% (209) | hypothetical protein MAV_3690 |
| M. smegmatis MC2 155 | MSMEG_1136 | - | 3e-06 | 26.47% (204) | hypothetical protein MSMEG_1136 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4369 | - | 4e-08 | 27.88% (165) | hypothetical protein Mvan_4369 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0957|M.abscessus_ATCC_1997 --MCAALGVGLLSGG-----AAVAHADP-------AFPDLSGFAVVPPDG
MAV_3690|M.avium_104 --MRAVVGVTRVLGGLAL--AAIAAVAPRAVAQPPGFPNLDGFTAVPSDG
Mflv_3413|M.gilvum_PYR-GCK --MTAWVRASVLAAG-VA--GAVVGASPSTAAPPPDFPDLAAYPAVDPAA
MSMEG_1136|M.smegmatis_MC2_155 MLSRLLVGLASAAGAAVMSVPGVAAADPQADPPPPTVPDVNAYPPVKTSE
Mvan_4369|M.vanbaalenii_PYR-1 MRSVALLVLFCFLAS-----ASVVSAQAE----PPGIPDLRGYRTVPVDD
: .. .:. . . .*:: .: *
MAB_0957|M.abscessus_ATCC_1997 FFVTNENSSVRSIHFSTPDGIGCMFRASPNMTPTSRQRLSCDGAVPGIPG
MAV_3690|M.avium_104 YLTSGAPGSPPRIGFSTPYAVACDFYGGPAPVPQPSQDIKCKGDMSGMD-
Mflv_3413|M.gilvum_PYR-GCK YQVTGGHPSTSGWAFTTAGGLRCQNSLIPDLG------IFCEGPVPDAP-
MSMEG_1136|M.smegmatis_MC2_155 YAVMDNN----WYAFSIGEGITCVLQRTGSYG--------CSGPIPAAP-
Mvan_4369|M.vanbaalenii_PYR-1 FVVDGAA------YFQTPDGLDCAILPTNGTAG-------CDGPLPATPA
: . . * .: * *.* :.
MAB_0957|M.abscessus_ATCC_1997 DAPNAPNVPGIGGA------SANVPGLGGVNIPGVGT------------C
MAV_3690|M.avium_104 ----------------------DVPFPGGR--PRPGD------------C
Mflv_3413|M.gilvum_PYR-GCK --------------------SEVMSVNVSLTTPAGFD------------R
MSMEG_1136|M.smegmatis_MC2_155 --------------------NGANLVSGGPGVPGFAS------------S
Mvan_4369|M.vanbaalenii_PYR-1 GANEIVLAAEVDTRGLRTTANPSFLAPPGHAVPELPEGAKIVYGDFECAA
. *
MAB_0957|M.abscessus_ATCC_1997 PMGTVAQKGDGAFEISKGAWSCGTKPEAPL--------------------
MAV_3690|M.avium_104 VLGTVDFKGPG-YQLSRMSYGGCDGNPAPLPSGGKT--------------
Mflv_3413|M.gilvum_PYR-GCK REGELETPWSSLLPVG-SRFAAGNGVVCAAP-------------------
MSMEG_1136|M.smegmatis_MC2_155 PAPVFAVVGDAKPLPPNTRLSYQTISCGFDG-------------------
Mvan_4369|M.vanbaalenii_PYR-1 GSGPITLCAKGTPAMQWMMISAERTGIGPATDGLPPGFPDPNDFVVGDDE
. . .
MAB_0957|M.abscessus_ATCC_1997 ----------------------------------------LNVGQKLTYG
MAV_3690|M.avium_104 ----------------------------------------LGAGQKLTYL
Mflv_3413|M.gilvum_PYR-GCK --------------------------------------------DEVTFA
MSMEG_1136|M.smegmatis_MC2_155 ----------------------------------------------VTTA
Mvan_4369|M.vanbaalenii_PYR-1 YLVGTGPKNMFPIFTVEGGLTCSIVTFSGGEIGCNGPLPGVSGGENEIFA
MAB_0957|M.abscessus_ATCC_1997 NVTCAVGAGSVTACQVVTGDQ-----------------------------
MAV_3690|M.avium_104 NVTCAVGADHLIACLDTTSG------------------------------
Mflv_3413|M.gilvum_PYR-GCK CWAAKPDSWPADTPDPPDRHYG----------------------------
MSMEG_1136|M.smegmatis_MC2_155 CIDGRNQAGFVISPTGSFVIG-----------------------------
Mvan_4369|M.vanbaalenii_PYR-1 QLPGATGIRRTDNPKFSTPDYPGQIRQLPVGHRVNGIGGTCMAIAGGVAC
MAB_0957|M.abscessus_ATCC_1997 -------KHGFVLQPSGSTSF-
MAV_3690|M.avium_104 -------DHGFVVQRTGSWAF-
Mflv_3413|M.gilvum_PYR-GCK -------EHGFVLSPAGNRGY-
MSMEG_1136|M.smegmatis_MC2_155 -------QQESDFKPTFQIG--
Mvan_4369|M.vanbaalenii_PYR-1 YGTVAGRAQGFEVSATETTTFG
: .. :