For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTEYQTLCVTTEDDRVIVELHRPEQRNAINAAMVADLHAVCATIEAQPRVLIVTGAGRDFAAGADIAELR VRGRNEALAGINRTVFDRIAALPLPTIAAVEGNALGGGAELAYACDIRIAGAGARFGNPEPGLGILAAAG ASYRLADLVGKSVAKQVILGGRVLDAEAALACGLVAEVVAEHSALGAASALTERITRQSPLALRLSKAVL DADSAHPLIDDLAQAVLFESPDKTDRMTRFLERQTR
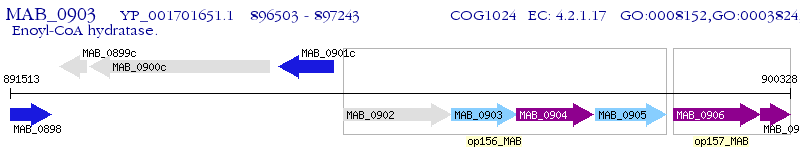
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0903 | - | - | 100% (246) | putative enoyl-CoA hydratase/isomerase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 6e-34 | 34.13% (252) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_0834 | - | 6e-29 | 36.44% (236) | putative enoyl-CoA hydratase/isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 3e-34 | 34.68% (248) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 8e-37 | 37.75% (249) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 3e-34 | 34.68% (248) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-33 | 34.54% (249) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4396 | echA8 | 9e-35 | 35.08% (248) | enoyl-CoA hydratase EchA8 |
| M. avium 104 | MAV_1195 | - | 2e-34 | 35.08% (248) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5277 | - | 2e-34 | 35.46% (251) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_4102 | - | 5e-35 | 36.90% (252) | PUTATIVE short chain enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 6e-35 | 35.08% (248) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4681 | - | 8e-33 | 35.34% (249) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4396|M.marinum_M ------MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELD
MUL_0210|M.ulcerans_Agy99 ------MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELD
MLBr_02402|M.leprae_Br4923 ------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELD
Mb1099c|M.bovis_AF2122/97 ------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELD
Rv1070c|M.tuberculosis_H37Rv ------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELD
MAV_1195|M.avium_104 MTDSTDRTFETILVEREERVGIITLNRPKALNALNTQVMNEVTTAATDFD
TH_4102|M.thermoresistible__bu ---MSAKNYETILVTREDRVGIITLNRPKALNALNSQVMTEVTDAAAEFD
Mflv_2036|M.gilvum_PYR-GCK ------MTFETILVDRDDRVATITLNRPKALNALNTQVMNEVTTAAAELD
Mvan_4681|M.vanbaalenii_PYR-1 ---MGSQNFETIIVERDDRVATITLNRPKALNALNSQVMDEVTTAAAELD
MSMEG_5277|M.smegmatis_MC2_155 -----MSDFETILVTRTDRVATIQLNRPKALNALNSQVMTEVTAAAAELD
MAB_0903|M.abscessus_ATCC_1997 -----MTEYQTLCVTTEDDRVIVELHRPEQRNAINAAMVADLHAVCATIE
::*: * : : *:**: **:*: :: :: .. ::
MMAR_4396|M.marinum_M NDPGIGAIIITGSAKAFAAGADIKEMADLTFSEAFDADFFAAWGKLAAVR
MUL_0210|M.ulcerans_Agy99 NDPGIGAIIITGSAKAFAAGADIKEMADLTFSEAFDADFFAAWGKLAAVR
MLBr_02402|M.leprae_Br4923 IDPDVGAILITGSPKVFAAGADIKEMASLTFTDAFDADFFSAWGKLAAVR
Mb1099c|M.bovis_AF2122/97 DDPDIGAIIITGSAKAFAAGADIKEMADLTFADAFTADFFATWGKLAAVR
Rv1070c|M.tuberculosis_H37Rv DDPDIGAIIITGSAKAFAAGADIKEMADLTFADAFTADFFATWGKLAAVR
MAV_1195|M.avium_104 ADPGIGAIIITGSAKAFAAGADIKEMAELTFADAYGADFFAPWGKLAALR
TH_4102|M.thermoresistible__bu SDPGIGAIIVTGNEKAFAAGADIKEMADLEFADVFAADFFAPWSKFAATR
Mflv_2036|M.gilvum_PYR-GCK AEPGVGAIIVTGSERAFAAGADIKEMASLSFADVFSSDFFAAWGKFAATR
Mvan_4681|M.vanbaalenii_PYR-1 ADAGVGAIILTGSEKAFAAGADIKEMAGLSFADVFSSDFFAGWGKFAATR
MSMEG_5277|M.smegmatis_MC2_155 KDPGVGAIIVTGNEKAFAAGADIKEMADLSFAEVFEADFFELWSKFAATR
MAB_0903|M.abscessus_ATCC_1997 AQPRV--LIVTGAGRDFAAGADIAELRVRGRNEALAGINRTVFDRIAALP
:. : :::** : ******* *: :. . :.::**
MMAR_4396|M.marinum_M TPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQ
MUL_0210|M.ulcerans_Agy99 TPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQ
MLBr_02402|M.leprae_Br4923 TPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQ
Mb1099c|M.bovis_AF2122/97 TPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
Rv1070c|M.tuberculosis_H37Rv TPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
MAV_1195|M.avium_104 TPTIAAVAGHALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
TH_4102|M.thermoresistible__bu TPTIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQ
Mflv_2036|M.gilvum_PYR-GCK TPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQ
Mvan_4681|M.vanbaalenii_PYR-1 TPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQ
MSMEG_5277|M.smegmatis_MC2_155 TPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQ
MAB_0903|M.abscessus_ATCC_1997 LPTIAAVEGNALGGGAELAYACDIRIAGAGARFGNPEPGLGILAAAGASY
* **** * *****.*:* **: **. *:**:** **:*.. *.*
MMAR_4396|M.marinum_M RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKAVAT
MUL_0210|M.ulcerans_Agy99 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKAVAT
MLBr_02402|M.leprae_Br4923 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVAT
Mb1099c|M.bovis_AF2122/97 RLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTEARATAT
Rv1070c|M.tuberculosis_H37Rv RLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTEARATAT
MAV_1195|M.avium_104 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKKVAT
TH_4102|M.thermoresistible__bu RLTRAIGKAKAMDLILTGRTIDAEEAERSGLVSRVVPADKLLDEAKETAA
Mflv_2036|M.gilvum_PYR-GCK RLTRAIGKAKAMDLILTGRNMDAEEAERAGLVSRVVPADSLLSEAKAVAQ
Mvan_4681|M.vanbaalenii_PYR-1 RLTRAIGKAKAMDLILTGRNMGAEEAERAGLVSRVVPADTLLEEARSVAK
MSMEG_5277|M.smegmatis_MC2_155 RLTRAIGKAKAMDLILTGRTIDAAEAERAGLVSRLVPADTLIDEALAVAQ
MAB_0903|M.abscessus_ATCC_1997 RLADLVGKSVAKQVILGGRVLDAEAALACGLVAEVVAEHSALGAASALTE
**: :**: * ::** ** :.* * .***:.:* . : * :
MMAR_4396|M.marinum_M TISQMSRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFATADQSEGMA
MUL_0210|M.ulcerans_Agy99 TISQMSRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFATADQSEGMA
MLBr_02402|M.leprae_Br4923 TISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMA
Mb1099c|M.bovis_AF2122/97 TISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMA
Rv1070c|M.tuberculosis_H37Rv TISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMA
MAV_1195|M.avium_104 TISQMSRSAARMAKEAVNRAFETTLAEGLLYERRLFHSTFATADQSEGMA
TH_4102|M.thermoresistible__bu TIAAMSLSAARMAKEAVNRAFETSLAEGLLFERRVFHSAFATADQSEGMA
Mflv_2036|M.gilvum_PYR-GCK TIAGMSLSASRMAKEAVDRAFETTLSEGLLYERRLFHSAFATDDQTEGMN
Mvan_4681|M.vanbaalenii_PYR-1 TIAGMSLSASRMAKEAVDRAFETTLTEGLLYERRLFHSAFATDDQTEGMN
MSMEG_5277|M.smegmatis_MC2_155 TIAGMSLSASRMAKEAVNRAFETSLTEGLLYERRLFHSAFATADQKEGMA
MAB_0903|M.abscessus_ATCC_1997 RITRQSPLALRLSKAVLDADSAHPLIDDLAQA-----VLFESPDKTDRMT
*: * * *::* .:: .* :.* * : *:.: *
MMAR_4396|M.marinum_M AFIEKRPPNFTHR
MUL_0210|M.ulcerans_Agy99 AFIEKRPPNFTHR
MLBr_02402|M.leprae_Br4923 AFIEKRAPQFTHR
Mb1099c|M.bovis_AF2122/97 AFIEKRAPQFTHR
Rv1070c|M.tuberculosis_H37Rv AFIEKRAPQFTHR
MAV_1195|M.avium_104 AFVEKRAPNFTHR
TH_4102|M.thermoresistible__bu AFTEKRPPKFTHR
Mflv_2036|M.gilvum_PYR-GCK AFSEKRSPNFTHR
Mvan_4681|M.vanbaalenii_PYR-1 AFTEKRSPNFTHR
MSMEG_5277|M.smegmatis_MC2_155 AFAEKRAANFTHR
MAB_0903|M.abscessus_ATCC_1997 RFLERQTR-----
* *::.