For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFAKTGSLIAGMVVLSVAGCSSTEPSGTGATTVRTSVASGPVDFAKIPGQIPAEAEMTQQGSEEAPIGAC VYLKGKPGSVTLNKVDCDSQDANYRVIQRVGFPDQCVNDADRRFYLGSPQGEWTACMDYAWTSEGCISVA PDKVVRAECDDKNLPNRERPITILFNTIDTSRCLFGGFAHPVRRFTVCTETQK
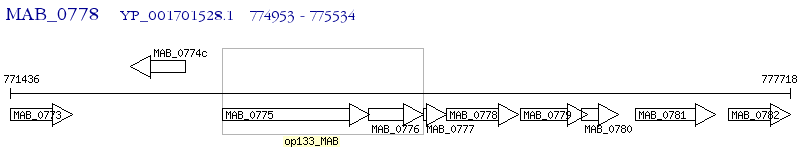
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0778 | - | - | 100% (193) | hypothetical protein MAB_0778 |
| M. abscessus ATCC 19977 | MAB_2981c | - | 2e-37 | 39.04% (187) | putative liporotein LppU |
| M. abscessus ATCC 19977 | MAB_3107c | - | 1e-13 | 31.62% (136) | lipoprotein LppU |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2807c | lppU | 3e-13 | 32.86% (140) | lipoprotein LppU |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2784c | lppU | 4e-13 | 32.86% (140) | lipoprotein LppU |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1923 | lppU | 2e-11 | 30.88% (136) | lipoprotein LppU |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2655 | - | 3e-15 | 33.33% (129) | LppU protein |
| M. thermoresistible (build 8) | TH_1282 | lppU | 9e-14 | 29.63% (135) | PROBABLE LIPOPROTEIN LPPU |
| M. ulcerans Agy99 | MUL_2148 | lppU | 1e-11 | 30.88% (136) | lipoprotein LppU |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2655|M.smegmatis_MC2_155 --------------------------------------------------
TH_1282|M.thermoresistible__bu -------------------------------MRTPPRDSPVAAGSAILAI
Mb2807c|M.bovis_AF2122/97 -------------------------------MR--AWLAAATTALFVVAT
Rv2784c|M.tuberculosis_H37Rv -------------------------------MR--AWLAAATTALFVVAT
MMAR_1923|M.marinum_M -------------------------------MR--ALFLAVLMALAVPAS
MUL_2148|M.ulcerans_Agy99 -------------------------------MR--ALFLAVLMALAVPAS
MAB_0778|M.abscessus_ATCC_1997 MFAKTGSLIAGMVVLSVAGCSSTEPSGTGATTVRTSVASGPVDFAKIPGQ
MSMEG_2655|M.smegmatis_MC2_155 --------------MQAGDCLELGGTFDQPEATRAECGSKKSNYKVVQTV
TH_1282|M.thermoresistible__bu VAVVGAVSGCASADLQVGDCLRMGGPPDNPDAAEVPCGTPESTHKIAAIV
Mb2807c|M.bovis_AF2122/97 ----GCSSATNVAELKVGDCVKLAGTPDRPQATKAECGSPASNFKVVAVV
Rv2784c|M.tuberculosis_H37Rv ----GCSSATNVAELKVGDCVKLAGTPDRPQATKAECGSPASNFKVVAVV
MMAR_1923|M.marinum_M GVLVGCSSTTKAADLAVGDCLKLAGPPDRPQATKAACGSEDSNFKVVAVA
MUL_2148|M.ulcerans_Agy99 GVLVGCSSTTKAADLAVGDCLKLAGPPDRPQATKAACGSEDSNFKVVAVA
MAB_0778|M.abscessus_ATCC_1997 IPAEAEMTQQGSEEAPIGACVYLKGKPGSVTLNKVDCDSQDANYRVIQRV
* *: : * . .. *.: :..:: .
MSMEG_2655|M.smegmatis_MC2_155 AD---SARCPADVDSYYTLSSRFGGETHTVCMDIDWVVGGCMNVDPENNT
TH_1282|M.thermoresistible__bu QD---EAECPADVDSYYSMRSTFDDATTTVCLDIDWVVGGCMSIDPTDEA
Mb2807c|M.bovis_AF2122/97 QE--DHAECPADVDSTYSMRNAFNGSTNTICLDIDWVIGGCMSVDPTHNT
Rv2784c|M.tuberculosis_H37Rv QE--DHAECPADVDSTYSMRNAFNGSTNTICLDIDWVIGGCMSVDPTHNT
MMAR_1923|M.marinum_M KDGTDHTECPADVDSSYSSRNVLGGANSTLCLDVDWVLGSCMSVDPDHKT
MUL_2148|M.ulcerans_Agy99 KDGTDHTECPADVDSSYSSRNVLGGANSTLCLDVDWVLGSCMSVDPDHKT
MAB_0778|M.abscessus_ATCC_1997 GFP---DQCVNDADRRFYLGSPQG--EWTACMDYAWTSEGCISVAP---D
.* *.* : . . * *:* *. .*:.: *
MSMEG_2655|M.smegmatis_MC2_155 DPYRVDCSDAGAPHRQRVTEVLEGISN---PDQCATGLGYAYDERQFTVC
TH_1282|M.thermoresistible__bu DPVRADCADPAAPHLQRVTEILDDVAD---VDQCVSGTGYAYEDRQFTVC
Mb2807c|M.bovis_AF2122/97 DPFRVDCDDASVPHRQRATQILKDLDSPVSVDQCASGVGYVYTQRRFAVC
Rv2784c|M.tuberculosis_H37Rv DPFRVDCDDASVPHRQRATQILKDLDSPVSVDQCASGVGYVYTQRRFAVC
MMAR_1923|M.marinum_M DPFRVGCNDASAPHRQRATQILQDVASPVTVDQCASGVGYTYTERRFVVC
MUL_2148|M.ulcerans_Agy99 DPFRVGCNDASAPHRQRATQILQDVASPVTVDQCASGVGYTYTERRFVVC
MAB_0778|M.abscessus_ATCC_1997 KVVRAECDDKNLPNRERPITILFNTID---TSRCLFG-GFAHPVRRFTVC
. *. * * *: :* :* . . .:* * *:.: *:*.**
MSMEG_2655|M.smegmatis_MC2_155 VENVR-----
TH_1282|M.thermoresistible__bu VEHVEV----
Mb2807c|M.bovis_AF2122/97 VEDVTGGPRS
Rv2784c|M.tuberculosis_H37Rv VEDVTGGPRS
MMAR_1923|M.marinum_M VEDVGGSSQT
MUL_2148|M.ulcerans_Agy99 VEDVGGSSQT
MAB_0778|M.abscessus_ATCC_1997 TETQK-----
.*