For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDTATAAPARSRWLPLQFAALALVWGASFLFIKVGLQGLSALQVAAARLDFGALTLVSLLVLLRVPLPRE IKVWGHIAVVSLTLCVMPFVLYPWAEQTIDSGLASIYNAATPLMTTLVTLLALRAERPDRTQLLGLALGF LGVCVVLAPWQLIGHGGPVLAQFACLAATLSYGIGFVYMRKYVTPLRLPALTVAAIQVGLGAVLMTAVVA LVDPHRIVATSAVVLSMLALGMLGTGLAYVWNTHIINGWGATAASSVTYLTPVIGVLLGALLLGEHIGWH EPLGGAIVIAGIVVSRRGSR
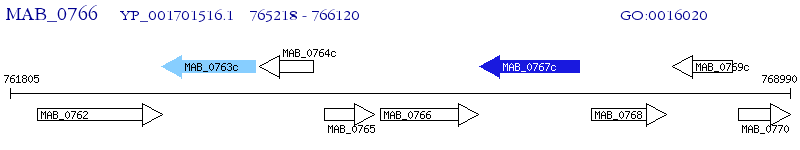
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0766 | - | - | 100% (300) | integral membrane protein |
| M. abscessus ATCC 19977 | MAB_0677c | - | 3e-14 | 24.74% (291) | hypothetical protein MAB_0677c |
| M. abscessus ATCC 19977 | MAB_3180 | - | 6e-10 | 23.16% (285) | hypothetical protein MAB_3180 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4964 | - | 3e-11 | 27.10% (310) | hypothetical protein Mflv_4964 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4903 | - | 2e-05 | 22.79% (215) | hypothetical protein MMAR_4903 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2188 | - | 2e-22 | 26.43% (280) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0035 | - | 1e-07 | 25.42% (299) | integral membrane protein |
| M. ulcerans Agy99 | MUL_0503 | - | 5e-06 | 23.26% (215) | hypothetical protein MUL_0503 |
| M. vanbaalenii PYR-1 | Mvan_5539 | - | 5e-23 | 28.01% (282) | hypothetical protein Mvan_5539 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4964|M.gilvum_PYR-GCK ---MAALDSSVTSRTQARADHFRSGLLFALASAFAFGCSGPFAKALMTAG
TH_0035|M.thermoresistible__bu ---LARIHSAVN--------HFRLGLVFAIASASAFGMSGPLAKSLLEAG
MSMEG_2188|M.smegmatis_MC2_155 --------------------------MLFGAMSVIWGIPYLLIKVAVEGV
Mvan_5539|M.vanbaalenii_PYR-1 --------------------MSLRGWLLFTAMGVIWGIPYLLIKVAVEGL
MMAR_4903|M.marinum_M -------------MPALAMGVTVTLWAFAFVGVRSAGRVFSPGVLALSRL
MUL_0503|M.ulcerans_Agy99 MASSTTSSQRTVNMPALAMGVTVTLWAFAFVGVRSAGRVFSPGVLALSRL
MAB_0766|M.abscessus_ATCC_1997 -----------MDTATAAPARSRWLPLQFAALALVWGASFLFIKVGLQGL
. * :
Mflv_4964|M.gilvum_PYR-GCK WSPTAAVTARLAGGALAMAVFATVVRPGWVREARHHARTVVLYGIVPIAG
TH_0035|M.thermoresistible__bu WSPAAAVTARLTGGALVLALFATAIKPGWLREAVRHRHTVVLYGIVPVAG
MSMEG_2188|M.smegmatis_MC2_155 SVPVLVLARTAVG---ALVLLPLAMSRAAWAPVLRHWKPVLVFAFFEIIA
Mvan_5539|M.vanbaalenii_PYR-1 SVPVLVFARTAVG---ALVLIPLTLRRGAWAPVLAHWRPVVAFAFFEIIA
MMAR_4903|M.marinum_M FIAVMVMAVIATIAATYTAYAGRATTRRLRPPSAAALTLVIAYGVAWFGA
MUL_0503|M.ulcerans_Agy99 FIAVMVMAVIATIAATYTAYAGRATTRRLRPPSAAALTLVIAYGVAWFGA
MAB_0766|M.abscessus_ATCC_1997 SALQVAAARLDFG--ALTLVSLLVLLRVPLPREIKVWGHIAVVSLTLCVM
. : . : ..
Mflv_4964|M.gilvum_PYR-GCK AQLFYFNAVAHLSVGVALLLEYTAPLLVVGWLWATTRHRPSGMT-LAGVV
TH_0035|M.thermoresistible__bu TQLAYYHAVSHLSVGVALLLEYTAPILVVGWLWLVHRRTPARLT-LAGTA
MSMEG_2188|M.smegmatis_MC2_155 AWALLTDAERHLTSSMTGLLIAAAPIIAAVLDRITGGER-LGVKRVAGLF
Mvan_5539|M.vanbaalenii_PYR-1 AWLLLSDAERHITSSLTGLLIAAAPIVAALLDRLTGGDQPLTVKRLAGLG
MMAR_4903|M.marinum_M YSIVISWAEQHIDAGTTALLVNLAPILVAVFAGFFLGEG-YSARLLTGIA
MUL_0503|M.ulcerans_Agy99 YSIVISWAEQHIDAGTTALLVNLAPILVAVFAGFFLGEG-YSARLLTGIA
MAB_0766|M.abscessus_ATCC_1997 PFVLYPWAEQTIDSGLASIYNAATPLMTTLVTLLALRAERPDRTQLLGLA
* : . : : :*::.. : *
Mflv_4964|M.gilvum_PYR-GCK LAVGGITLVLGIIGPGGVAAAHINLAGVGWG-LAAAVCAACYFLMSDKAG
TH_0035|M.thermoresistible__bu LAVGGMTLVLDVFS-----GAHFDVVGLAWG-LTAAVCSASYFLMSDRVG
MSMEG_2188|M.smegmatis_MC2_155 VGLSGVAVLAGPGLAG----------GHVWP-VLEVLLVATCYAIAPLVA
Mvan_5539|M.vanbaalenii_PYR-1 TGLAGVAVLAGPELAG----------GSAWP-VTEVLLVAVCYAIAPLIA
MMAR_4903|M.marinum_M IAFAGVVLISAGGGGS----------QTDWLGVGLGLLAAVLYAAGVLLQ
MUL_0503|M.ulcerans_Agy99 IAFAGVVLISAGGGGS----------QTDWLGVGLGLLAAVLYAAGVLLQ
MAB_0766|M.abscessus_ATCC_1997 LGFLGVCVVLAPWQLIG--------HGGPVLAQFACLAATLSYGIGFVYM
.. *: :: : : : .
Mflv_4964|M.gilvum_PYR-GCK AMSDKEVADGRPSLHPITLAAAGLAFGAVTVAALGFSGLMPLAFTANDAV
TH_0035|M.thermoresistible__bu TGED--------GLHPVTLAAGGLLVGAVTVATAGAVGIAPLTFTTGDTV
MSMEG_2188|M.smegmatis_MC2_155 ARFLG--------------EVPSLPMTAACLTVAALIYTTPAALTWPDAM
Mvan_5539|M.vanbaalenii_PYR-1 ARYLA--------------EVPAMPLTAACLGLAALIYVGPAAATWPAEI
MMAR_4903|M.marinum_M KVALR--------------TVDALSATLWGTLVGFIVTLPFLPAAARELS
MUL_0503|M.ulcerans_Agy99 KVALR--------------TVDALSATLWGTLVGFIVTLPFLPAAARELS
MAB_0766|M.abscessus_ATCC_1997 RKYVTP------------LRLPALTVAAIQVGLGAVLMTAVVALVDPHRI
.: . .
Mflv_4964|M.gilvum_PYR-GCK IAGFTTPWIVPVIALALIPTAIAYTLGIVGIARLRPRFASMVGLSEVMFA
TH_0035|M.thermoresistible__bu IAGVTMSWLVPVIGLGVIATAIAYTLGIIGIALLRPRFASLVGLSEVMFA
MSMEG_2188|M.smegmatis_MC2_155 PS---TRVLAALAALAVVCTALAFIVFFALIREVGAARALVFTYVNPAVA
Mvan_5539|M.vanbaalenii_PYR-1 PS---MRVLTAVALLAVVCTSLAFILFFALIREVGAPRALVITYVNPAVA
MMAR_4903|M.marinum_M LAS--AADLWWLVFLGAGPSAIAFTTWAYALARTNAGVTAATTLMVPALV
MUL_0503|M.ulcerans_Agy99 LAS--AADLWWLVFLGAGPSAIAFTTWAYALARTNAGVTAATTLMVPALV
MAB_0766|M.abscessus_ATCC_1997 VAT--SAVVLSMLALGMLGTGLAYVWNTHIINGWGATAASSVTYLTPVIG
: : : *. :.:*: : . : .
Mflv_4964|M.gilvum_PYR-GCK VLAAWMLLGEGMAAAQVIGGAIVLAGLALARQGDRG-ELVGETNVGETSV
TH_0035|M.thermoresistible__bu TVAAWLLIGETVTPVQAVGGVIVLTGLALARQGDR-------------PV
MSMEG_2188|M.smegmatis_MC2_155 LAAGVIVLGEPLTAYNIAGLVLILSGSVLATRKATHGNRERETSAKNQPD
Mvan_5539|M.vanbaalenii_PYR-1 LAAGVIVLNEPLTARHLLGLAMILAGSVLATRRP----------------
MMAR_4903|M.marinum_M IALSGLLLAEVPNPLGLAGGGACLIGVALARGLLRLPARSRRRTGTGDQL
MUL_0503|M.ulcerans_Agy99 IALSGLLLAEVPNPLGLAGGGACLIGVVLARGLLRLPARSRRRTGTGDQL
MAB_0766|M.abscessus_ATCC_1997 VLLGALLLGEHIGWHEPLGGAIVIAGIVVSRRGSR---------------
. ::: * * : * .::
Mflv_4964|M.gilvum_PYR-GCK TPTALPQTTPVARGGPATIDP-----------
TH_0035|M.thermoresistible__bu PGTGDTPTTPVDDGPDRRVPELH---------
MSMEG_2188|M.smegmatis_MC2_155 SRSQFTLAEPIEAIEPIEPKGPTGPGATRRTP
Mvan_5539|M.vanbaalenii_PYR-1 -------VEPMQAAPR----------------
MMAR_4903|M.marinum_M QGVESLPVGLSDYAASSTAPSGRQL-------
MUL_0503|M.ulcerans_Agy99 QGVESLPVGLSDYAASSTAPSGRQL-------
MAB_0766|M.abscessus_ATCC_1997 --------------------------------