For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLEGSLDLLLLTADPNPDGVLPSLSLLAHTVRPVPSEVSSLLEAGAADVAIVDARTDLASARGLCRLLGA AGSSVPVVAVVNEGSLVAVNVDWGLDDILLPGTGPAEIDARLRLLVGRRAGAVNVENASKVVLGELVIDE GTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLQEVWGYDFFGGTRTVDVHVRRLRAKLGGEYE SLIGTVRNVGYKAVRPPRAKGEPGPSAADDDDESDIDDLDDDGTDQDIASEPGAINAG
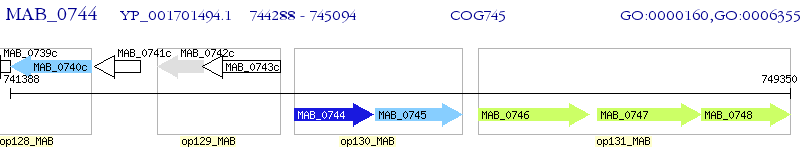
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0744 | - | - | 100% (268) | transcriptional regulatory protein |
| M. abscessus ATCC 19977 | MAB_4047c | - | 3e-25 | 35.00% (200) | sensory transduction protein RegX3 |
| M. abscessus ATCC 19977 | MAB_0956c | - | 5e-21 | 36.16% (177) | transcriptional regulatory protein PrrA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0841 | - | 1e-100 | 76.89% (238) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1650 | - | 1e-114 | 80.61% (263) | response regulator receiver protein |
| M. tuberculosis H37Rv | Rv0818 | - | 9e-99 | 76.47% (238) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02439 | - | 1e-22 | 35.14% (185) | putative two-component system response regulator |
| M. marinum M | MMAR_4865 | - | 1e-101 | 74.60% (252) | transcriptional regulatory protein |
| M. avium 104 | MAV_0760 | - | 1e-105 | 76.47% (255) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_5784 | - | 1e-111 | 80.24% (253) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_4312 | - | 3e-25 | 37.10% (186) | PUTATIVE two component transcriptional regulator, winged helix |
| M. ulcerans Agy99 | MUL_0435 | - | 1e-101 | 74.40% (250) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5097 | - | 1e-110 | 81.45% (248) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1650|M.gilvum_PYR-GCK MRCPGERAPRLLEGQLDLLLLTVDPHPESVLPSLSLLAHAVRTAPTEVSS
Mvan_5097|M.vanbaalenii_PYR-1 ---------------MDLLLLTVDPHPESVLPSLALLAHNVRTAPTEVSS
MSMEG_5784|M.smegmatis_MC2_155 ---------------MDLLLLTVDPHPESVLPSLSLLAHTVRTAPTEVSS
MAB_0744|M.abscessus_ATCC_1997 ----------MLEGSLDLLLLTADPNPDGVLPSLSLLAHTVRPVPSEVSS
Mb0841|M.bovis_AF2122/97 --------------MLELLLLTSELYPDPVLPALSLLPHTVRTAPAEASS
Rv0818|M.tuberculosis_H37Rv --------------MLELLLLTSELYPDPVLPALSLLPHTVRTAPAEASS
MMAR_4865|M.marinum_M -----MPFPRRWRGLLELLLLTSELYPDPVLPSLSLLPHNVRTAPAEASS
MUL_0435|M.ulcerans_Agy99 --------------MLELLLLTSELYPDPVLPSLSLLPHNVRTAPAEASS
MAV_0760|M.avium_104 ---------------MELLLLTPELHPDPVLPSLSLLAHTVRTAPPEPSS
MLBr_02439|M.leprae_Br4923 ----------------------------------------MVTDGPAALA
TH_4312|M.thermoresistible__bu -----------MTSVLIVEDEESLADPLAFLLRKEGFETTIVTDGPSALA
: . . :
Mflv_1650|M.gilvum_PYR-GCK LLEAGTADVAIVDARTDLAAARGLCRLLGTTGTSVPVVAVVNEGG--LVA
Mvan_5097|M.vanbaalenii_PYR-1 LLEAGTADVAIVDARTDLAAARGLCRLLGTTGTSVPVVAVVNEGG--LVA
MSMEG_5784|M.smegmatis_MC2_155 LLETGSADVAIVDARTDLAAARGLCRLLGTTGTSVPVVAVINEGG--LVA
MAB_0744|M.abscessus_ATCC_1997 LLEAGAADVAIVDARTDLASARGLCRLLGAAGSSVPVVAVVNEGS--LVA
Mb0841|M.bovis_AF2122/97 LLEAGNADAVLVDARNDLSSGRGLCRLLSSTGRSIPVLAVVSEGG--LVA
Rv0818|M.tuberculosis_H37Rv LLEAGNADAVLVDARNDLSSGRGLCRLLSSTGRSIPVLAVVSEGG--LVA
MMAR_4865|M.marinum_M LLEAGNADAVLVDARNDLSAARGLCRLLSTAGRSVPVLAVVNEGG--LVA
MUL_0435|M.ulcerans_Agy99 LLEAGNADAVLADARNDLSAARGLCRLLSTAGRSVPVLAVVNEGG--LVA
MAV_0760|M.avium_104 LLEAGTADAVIVDARTDLSSARGLCRLLSTAGRSVPVLAVVSEGG--LVA
MLBr_02439|M.leprae_Br4923 EFDRSGADIVLLDLMLPGMSGTDVYKQLRVR-SSVPVIMVTARDSEIDKV
TH_4312|M.thermoresistible__bu EFERSGADIILLDLMLPGMSGTDVCRRLRAK-SKVPVIMVTARDSEIDKV
:: . ** : * :. .: : * .:**: * ... .
Mflv_1650|M.gilvum_PYR-GCK VNVEWGLDEILLPSTGPAEIDARLRLLVGRRGGMANQENLG-KISLGELV
Mvan_5097|M.vanbaalenii_PYR-1 VNVEWGLDEILLPSTGPAEIDARLRLLVGRRGGMANQENLG-KISLGELV
MSMEG_5784|M.smegmatis_MC2_155 VNHEWGLDEILLPSTGPAEIDARLRLLVGRRGGNANQENVG-KITLGELV
MAB_0744|M.abscessus_ATCC_1997 VNVDWGLDDILLPGTGPAEIDARLRLLVGRRAGAVNVENAS-KVVLGELV
Mb0841|M.bovis_AF2122/97 VSADWGLDEILLPSTGPAEIDARLRLVVGRRGDLADQESLG-KVSLGELV
Rv0818|M.tuberculosis_H37Rv VSADWGLDEILLLSTGPAEIDARLRLVVGRRGDLADQESLG-KVSLGELV
MMAR_4865|M.marinum_M VSADWGLDEILLPSTGPAEIDARLRLVVGRRGGQADQESVG-KVSLGELV
MUL_0435|M.ulcerans_Agy99 VSADWGLDEILLPSTGPAEIDARLRLVVGRRGGQADQESVG-KVSLGELV
MAV_0760|M.avium_104 VNSDWGLDEILLPTTGPAEVDARLRLVVGRRGGLADQESAG-KVTLGELV
MLBr_02439|M.leprae_Br4923 VGLELGADDYVTKPYSARELIARIRAVLRRGGDDD-SEISDGVLESGPLR
TH_4312|M.thermoresistible__bu VGLELGADDYVTKPYSARELIARIRAVLRRGSDLDGADVDDSVLEAGPVR
*. : * *: : .. *: **:* :: * .. : . : * :
Mflv_1650|M.gilvum_PYR-GCK IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLQEVWGYDF
Mvan_5097|M.vanbaalenii_PYR-1 IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLQEVWGYDF
MSMEG_5784|M.smegmatis_MC2_155 IDEGTYTARLRGKPLDLTYKEFELLKYLAQHAGRVFTRAQLLQEVWGYDF
MAB_0744|M.abscessus_ATCC_1997 IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLQEVWGYDF
Mb0841|M.bovis_AF2122/97 IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLHEVWGYDF
Rv0818|M.tuberculosis_H37Rv IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLHEVWGYDF
MMAR_4865|M.marinum_M IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLHEVWGYDF
MUL_0435|M.ulcerans_Agy99 IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLHEVWGYDF
MAV_0760|M.avium_104 IDEGTYTARLRGRPLDLTYKEFELLKYLAQHAGRVFTRAQLLHEVWGYDF
MLBr_02439|M.leprae_Br4923 MDVERHVVSVNGYAITLPLKEFDLLEYLMRNSGRVLTRGQLIDRVWGVDY
TH_4312|M.thermoresistible__bu MDVDRHVVTVRGEPITLPLKEFELLEYLLRNSGRVLTRGQLIDRVWGADY
:* :.. :.* .: *. ***:**:** :::***:**.**:..*** *:
Mflv_1650|M.gilvum_PYR-GCK FGGTRTVDVHVRRLRAKLG--PEYESLIGTVRNVGYKAVRPARGR-PPAP
Mvan_5097|M.vanbaalenii_PYR-1 FGGTRTVDVHVRRLRAKLG--PEYEALIGTVRNVGYKAVRPARGR-APAA
MSMEG_5784|M.smegmatis_MC2_155 FGGTRTVDVHVRRLRAKLG--PEYEALIGTVRNVGYKAVRPSRGK-PPAA
MAB_0744|M.abscessus_ATCC_1997 FGGTRTVDVHVRRLRAKLG--GEYESLIGTVRNVGYKAVRPPRAKGEPGP
Mb0841|M.bovis_AF2122/97 FGGTRTVDVHVRRLRAKLG--PEHEALIGTVRNVGYKAVRPARGR-PPAA
Rv0818|M.tuberculosis_H37Rv FGGTRTVDVHVRRLRAKLG--PEHEALIGTVRNVGYKAVRPARGR-PPAA
MMAR_4865|M.marinum_M FGGTRTVDVHVRRLRAKLG--PEHEALIGTVRNVGYKAVRPARGR-SVAV
MUL_0435|M.ulcerans_Agy99 FGGTRTVDVHVRRLRAKLG--PEHEALIGTVRNVGYKAVRPARGR-SVAV
MAV_0760|M.avium_104 FGGTRTVDVHVRRLRAKLG--PEYEALIGTVRNVGYKAVRPARGR-PPVA
MLBr_02439|M.leprae_Br4923 VGDTKTLDVHVKRLRSKIEADPANPVHLVTVRGLGYKLES----------
TH_4312|M.thermoresistible__bu VGDTKTLDVHVKRLRSKIEEDPANPVHLVTVRGLGYKLEG----------
.*.*:*:****:***:*: : ***.:***
Mflv_1650|M.gilvum_PYR-GCK --ESEADDLD-DDPYADDE-PLASDEVPQALGDALRSP
Mvan_5097|M.vanbaalenii_PYR-1 GADLPSDDLDGDDPYEDGVGSLDRDGVPEALGDALRSQ
MSMEG_5784|M.smegmatis_MC2_155 --DASGEDVP-DGPDEGFG---DDVDVDGPLAGRLTSQ
MAB_0744|M.abscessus_ATCC_1997 SAADDDDESDIDDLDDDGT----DQDIASEPG-AINAG
Mb0841|M.bovis_AF2122/97 DPDDEDADPG-------------RDGMQEPLVDPLRSQ
Rv0818|M.tuberculosis_H37Rv DPDDEDADPG-------------RDGMQEPLVDPLRSQ
MMAR_4865|M.marinum_M EADDEEAESD-------------HDGYEEPLVDPLRSP
MUL_0435|M.ulcerans_Agy99 DADDEETESD-------------HDGYEEPLVDPLRSP
MAV_0760|M.avium_104 EADDAESESDSASES-------DAEDVHDPLVDPLHTQ
MLBr_02439|M.leprae_Br4923 --------------------------------------
TH_4312|M.thermoresistible__bu --------------------------------------