For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPQALVLLAAALLLVPQRYRRLDQPGMSPVRWTGKLVAAPLSALVLTALWLLPLSVVVSAVVVAGTGAL RWRRRRRSRVESEELDCTASGLDVVVGELRVGAHPVRAFEIAAVELNGAAGEAFAAIAARARLGVNVDRA LPRSVPARAQWNRITRSWSLAQRHGLAVADLLDACRADLQERQQFTARVNAGLAGPRATAVVLTGLPVAG IGLGQMIGARPLSVLCAGGVGGALLVVGVTLACAGLLWADAITGKALR
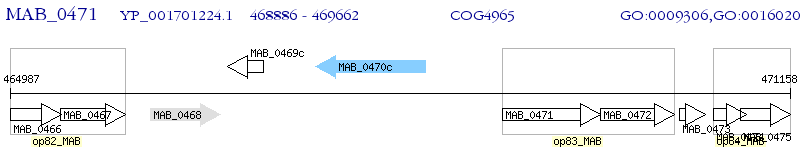
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0471 | - | - | 100% (258) | hypothetical protein MAB_0471 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3682c | - | 9e-56 | 48.64% (257) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1385 | - | 2e-46 | 43.46% (260) | type II secretion system protein |
| M. tuberculosis H37Rv | Rv3658c | - | 9e-56 | 48.64% (257) | transmembrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5146 | - | 3e-53 | 45.98% (261) | hypothetical protein MMAR_5146 |
| M. avium 104 | MAV_0509 | - | 3e-52 | 46.51% (258) | type II secretion system protein F |
| M. smegmatis MC2 155 | MSMEG_6168 | - | 5e-52 | 52.66% (207) | type II secretion system protein F |
| M. thermoresistible (build 8) | TH_3280 | - | 5e-35 | 50.33% (151) | PUTATIVE type II secretion system protein |
| M. ulcerans Agy99 | MUL_4231 | - | 1e-45 | 45.19% (239) | hypothetical protein MUL_4231 |
| M. vanbaalenii PYR-1 | Mvan_5406 | - | 1e-47 | 46.27% (255) | type II secretion system protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1385|M.gilvum_PYR-GCK ---MTLAALALAVAVLTAPTDARLRAPVLRRTSTP----RRRLPGQLPAA
Mvan_5406|M.vanbaalenii_PYR-1 ---MTAAALALAAAVLVAPAGSRWRR-VLSAPPSR----RLRLPVPVCGA
TH_3280|M.thermoresistible__bu --------------------------------------------------
Mb3682c|M.bovis_AF2122/97 MSGIASAALILSLALVVLPGSPRCRLTPDDTGRR----VLLVGARRVAWG
Rv3658c|M.tuberculosis_H37Rv MSGIASAALILSLALVVLPGSPRCRLTPDDTGRR----VLLVGARRVAWG
MAV_0509|M.avium_104 MTGPVVGALLLAVALVVGPPSPRRRLGVAVTGSRRPSARRLAGLGWVAAG
MMAR_5146|M.marinum_M MTGLVLAALMVAAAIAVLSVPVRRRISVNDVRGHS---NWLSATRWLGCL
MUL_4231|M.ulcerans_Agy99 MTGLVLAALMVAAAIAVLSVPVRRRISVNDVRGHS---NWLSATRWLGCL
MSMEG_6168|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0471|M.abscessus_ATCC_1997 ---MTPQALVLLAAALLLVPQRYRRLDQPGMSPVR------WTGKLVAAP
Mflv_1385|M.gilvum_PYR-GCK IACLTV--AAVHTPTTAAALGLLLATLSARRRVTRAHRRRTAEAAALQDA
Mvan_5406|M.vanbaalenii_PYR-1 LLCAVT--ALLLSPAAVLAAALLMATLSVRSRAARRRRNRVTEAAALQGA
TH_3280|M.thermoresistible__bu --------------------------------------------------
Mb3682c|M.bovis_AF2122/97 VGCVAVGVAALLPLPTVVAVAVLGATLGLRYRRRRRYLRRSREGQALEAA
Rv3658c|M.tuberculosis_H37Rv VGCVAVGVAALLPLPTVVAVAVLGATLGLRYRRRRRYLRRSREGQALEAA
MAV_0509|M.avium_104 AGCAAL---AVLPLPSVLAGAVVAATAGLRHRRRGRIRRAAGEGRALETA
MMAR_5146|M.marinum_M LACALAGAVLVMPLTTVLAAVVVAGTASRRRRRRLVRARGVREGRALEAA
MUL_4231|M.ulcerans_Agy99 LACALAGAVLVMPLTTVLAVAVVAGTASRRRRRRLVRARGVREGWALEAA
MSMEG_6168|M.smegmatis_MC2_155 ----------MLPLPVALTAGVVAGTVVVRRRDRAAAQHRRDESAALQGA
MAB_0471|M.abscessus_ATCC_1997 LSALVLTALWLLPLSVVVSAVVVAGTGALRWRRRRRSRVESEELDCTASG
Mflv_1385|M.gilvum_PYR-GCK LDVLVGELRVGAHPVAAITVAGHEARGRIAESLNAVAARALLGADVPAGL
Mvan_5406|M.vanbaalenii_PYR-1 LDVLIGELRVGAHPVAAMNTAAQESDARIAGALGAVAARALLGADVAAGL
TH_3280|M.thermoresistible__bu --------------VSAVEVAAREVSGTTGLALREVAARARLGGEVSSGL
Mb3682c|M.bovis_AF2122/97 LELVVGELRAGAHPVRAFSIAADETGGPVAVALRAVAARARLGADVTAGL
Rv3658c|M.tuberculosis_H37Rv LELVVGELRAGAHPVRAFSIAADETGGPVAVALRAVAARARLGADVTAGL
MAV_0509|M.avium_104 LDVLVGELRAGSHPVRAFDVAAGETAGPVAVALRSVAARARLGADVAAGL
MMAR_5146|M.marinum_M LEILVGELRVGAHPVRAFGVAADETDGPVAAAMRGVAAHARLGADVTGGL
MUL_4231|M.ulcerans_Agy99 LEILVGELRVGAHPVRAFGVAADETDGPVAAAMRGVAAHARLGADVTGGL
MSMEG_6168|M.smegmatis_MC2_155 LDVLVGELRVGAHPVDAFGAAAGESGGPVAEGMRAVAARARLGADVAAGL
MAB_0471|M.abscessus_ATCC_1997 LDVVVGELRVGAHPVRAFEIAAVELNGAAGEAFAAIAARARLGVNVDR--
* *. *. * . . .: :**:* ** :*
Mflv_1385|M.gilvum_PYR-GCK RAEGRRSPSPGHWDRLAVCWQLAQTHGLAIAALMRAAQRDITERERFRGR
Mvan_5406|M.vanbaalenii_PYR-1 RAQGRRSLLPGHWDRLAVCWHLAQAQGLAIATLMLAAQRDIAERERFRGR
TH_3280|M.thermoresistible__bu RSVAARSASPGNWERLAVCWRLAERYGLAMATLMRTAQHDIRERERFSAK
Mb3682c|M.bovis_AF2122/97 LAAARSSALPAYWERLAVCWQLGSDHGLAIASLMRAAQRDVAERQRFSAR
Rv3658c|M.tuberculosis_H37Rv LAAARSSALPAYWERLAVCWQLGSDHGLAIASLMRAAQRDVAERQRFSAR
MAV_0509|M.avium_104 AAASRGSARPEHWQRLALCWRLAGDHGLGIATLMRAAQRDIAERQRFSAR
MMAR_5146|M.marinum_M RAAARRSALPSHWERLAVYWQLASDHGLAIATLMRAAQRDVAQRQRFSAK
MUL_4231|M.ulcerans_Agy99 RAAARRSALPAHWERLAVYWQLASDHGLAIATLMRAAQRDVAQRQRFSAK
MSMEG_6168|M.smegmatis_MC2_155 DDVARASQLPGHWQRLAVCWRLAQTHGLALGTLMRAAHEDIVERERFSAR
MAB_0471|M.abscessus_ATCC_1997 -ALPRSVPARAQWNRITRSWSLAQRHGLAVADLLDACRADLQERQQFTAR
*:*:: * *. **.:. *: :.: *: :*::* .:
Mflv_1385|M.gilvum_PYR-GCK VESGMAGARATAVILAGLPVLGVLLGHAIGAEPLAFLLSGGAGGWLLLVG
Mvan_5406|M.vanbaalenii_PYR-1 VEAGLAGARATAAILAGLPVLGVLLGHAIGAEPLSFLFSGGFGGWLLVTG
TH_3280|M.thermoresistible__bu VHADLAGARTTATLLAALPVLGIGLGQAIGAEPVRFLCTDGIGAWLLVTG
Mb3682c|M.bovis_AF2122/97 VSAGMAGARASAAILAILPLLGVLLGQLIGARPLSFLLTGRVGGWLLVVG
Rv3658c|M.tuberculosis_H37Rv VSAGMAGARASAAILAILPLLGVLLGQLIGARPLSFLLTGRVGGWLLVVG
MAV_0509|M.avium_104 VSSSLAGARATAAILAALPVLGVLLGQLIGARPLDFLLGGQVGGWLLLVG
MMAR_5146|M.marinum_M VSAGLAGARASAGILAGLPVLGVVLGQLIGAQPLAFLLGGHLGGWLLVVG
MUL_4231|M.ulcerans_Agy99 VSAGLAGARASAGILAGLPVLGVVLGQLIGAQPLAFLLGGHLGGWLL--G
MSMEG_6168|M.smegmatis_MC2_155 VRAAMAGPRTTALVLAGLPLAGIALGQAIGAAPVAFLVASGAGGWLLVIG
MAB_0471|M.abscessus_ATCC_1997 VNAGLAGPRATAVVLTGLPVAGIGLGQMIGARPLSVLCAGGVGGALLVVG
* : :**.*::* :*: **: *: **: *** *: .* . *. ** *
Mflv_1385|M.gilvum_PYR-GCK TVFVCAGLLWSDRITVQVLT
Mvan_5406|M.vanbaalenii_PYR-1 TVFVCCGLLWSDRITTGVLT
TH_3280|M.thermoresistible__bu AALLCCGQLWSDRIIEGVVK
Mb3682c|M.bovis_AF2122/97 LTLACAGLLWSDRITDRPVL
Rv3658c|M.tuberculosis_H37Rv LTLACAGLLWSDRITDRPVL
MAV_0509|M.avium_104 ATLACAGLLWADRIVDRVMA
MMAR_5146|M.marinum_M LALAAGGLWWSDRITDRVMP
MUL_4231|M.ulcerans_Agy99 FAWPWRPVVCGGRTGSPTG-
MSMEG_6168|M.smegmatis_MC2_155 VVLACAGLLWSDRITAEVKS
MAB_0471|M.abscessus_ATCC_1997 VTLACAGLLWADAITGKALR
. ..