For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRYGLPVLIALFGLFAAAIPAAAAPAAGADDKVPMGGGAGLIINGDTLCTLTTIGHDGKGNLVGFTSAHC GGAGSQVASEDFPKKGVLGKFVNGNEQYDYATIQFDPEKVQPVSTYKGFTITGIGPDPQPGDIACKLGRT TGNSCGVTFGAGAEPGTFINQVCGQPGDSGAPVTVNGQLVGMIHGAATKLPVCVIKYIPLHTPTTTYSIN TVLADINAKNRTAGVGFTPVG
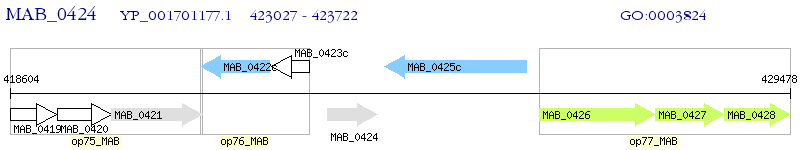
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0424 | - | - | 100% (231) | putative protease |
| M. abscessus ATCC 19977 | MAB_2805c | - | 1e-08 | 28.90% (173) | hypothetical protein MAB_2805c |
| M. abscessus ATCC 19977 | MAB_3348 | - | 7e-07 | 28.33% (180) | hypothetical protein MAB_3348 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3692c | - | 8e-79 | 62.61% (222) | putative protease |
| M. gilvum PYR-GCK | Mflv_1379 | - | 1e-76 | 67.50% (200) | peptidase S1 and S6, chymotrypsin/Hap |
| M. tuberculosis H37Rv | Rv3668c | - | 8e-79 | 62.61% (222) | protease |
| M. leprae Br4923 | MLBr_02295 | - | 2e-75 | 58.93% (224) | hypothetical protein MLBr_02295 |
| M. marinum M | MMAR_5156 | - | 9e-77 | 64.02% (214) | hypothetical protein MMAR_5156 |
| M. avium 104 | MAV_0461 | - | 4e-80 | 64.49% (214) | hypothetical protein MAV_0461 |
| M. smegmatis MC2 155 | MSMEG_6180 | - | 6e-76 | 60.53% (228) | secreted protein |
| M. thermoresistible (build 8) | TH_0965 | - | 2e-82 | 65.04% (226) | POSSIBLE PROTEASE |
| M. ulcerans Agy99 | MUL_4242 | - | 5e-76 | 63.55% (214) | hypothetical protein MUL_4242 |
| M. vanbaalenii PYR-1 | Mvan_5427 | - | 8e-75 | 59.65% (228) | peptidase S1 and S6, chymotrypsin/Hap |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02295|M.leprae_Br4923 --MVLRRGHWCILVALVAVLLAVVSMPAKTVFADGRLPMGGGAGIVINGD
MAV_0461|M.avium_104 -------------MAMVALLVAVTGSHPRAAAADVRIPLGGGAGIVVNGD
Mb3692c|M.bovis_AF2122/97 ----MQTAHRRFAAAFAAVLLAVVCLPANTAAADDKLPLGGGAGIVVNGD
Rv3668c|M.tuberculosis_H37Rv ----MQTAHRRFAAAFAAVLLAVVCLPANTAAADDKLPLGGGAGIVVNGD
MMAR_5156|M.marinum_M ----MQTAHRRISSAIVTVLLALVFGPANAAAADDKVPLGGGAGIVVNGG
MUL_4242|M.ulcerans_Agy99 ----MQTAHRRISSAIVTVLLALVFGPANAAAADDKVPLGGGAGIVVNGG
Mflv_1379|M.gilvum_PYR-GCK ------------------------------------MPLGGGSGLIVNGE
Mvan_5427|M.vanbaalenii_PYR-1 MRISGRSVPVRAAILALTALLTLTIVP---AHAATPVPLGGGSGLVVNGE
TH_0965|M.thermoresistible__bu ----------MLPIVALTGLLVAALVPSVVAHAAGPVPLGGGSGLVVNGE
MSMEG_6180|M.smegmatis_MC2_155 -------MLVGAPVVALFAVLAPWTSG--HAQAAPPVVLGGGSGIVVDGE
MAB_0424|M.abscessus_ATCC_1997 ----MRYGLPVLIALFGLFAAAIPAAAAPAAGADDKVPMGGGAGLIINGD
: :***:*::::*
MLBr_02295|M.leprae_Br4923 TMCTLTTIGTDSAAELIGFTSAHCGGPGAQVAAEGAENRGPMGTMIAGND
MAV_0461|M.avium_104 TMCTLTTIGGDAAGDLIGFTSAHCGGPGAQVAAEGAENAGILGTMVAGND
Mb3692c|M.bovis_AF2122/97 TMCTLTTIGHDKNGDLIGFTSAHCGGPGAQIAAEGAENAGPVGIMVAGND
Rv3668c|M.tuberculosis_H37Rv TMCTLTTIGHDKNGDLIGFTSAHCGGPGAQIAAEGAENAGPVGIMVAGND
MMAR_5156|M.marinum_M TLCTLTTIGHDKNGDLIGFSSAHCGGPGAPVAAEGAEDKGVVGTMVAGND
MUL_4242|M.ulcerans_Agy99 TLCTLTTIGHDKNGDLIGFSSAHCGGPGAPVAAEGAEDKGVVGTMVAGND
Mflv_1379|M.gilvum_PYR-GCK TLCTLTAIGTDNRGDLVGFTSAHCGGPGAVVASEAAPKAGVVGEMVAGND
Mvan_5427|M.vanbaalenii_PYR-1 TLCTLTAIGTDNQGSLIGFTSAHCGGPGAVVAAEGAENTGPVGTMVAGND
TH_0965|M.thermoresistible__bu TLCTLTTIGHDNRGALVGFTSAHCGGPGAVVSAEGAEDAGVLGTMVAGND
MSMEG_6180|M.smegmatis_MC2_155 SFCTLTAIGHDNAGRLIGFTSAHCGGPGATVAAESDVAAGVLGTMVAGND
MAB_0424|M.abscessus_ATCC_1997 TLCTLTTIGHDGKGNLVGFTSAHCGGAGSQVASEDFPKKGVLGKFVNGNE
::****:** * . *:**:******.*: :::* * :* :: **:
MLBr_02295|M.leprae_Br4923 NLDYAVIKFDPAKVMPVAAYNGFVISGIGQDPAFGQIACKQGRTTGNSCG
MAV_0461|M.avium_104 NLDYAVIKFDPAKVTPVANFNGFLISGIGPDPAFGEIACKQGRTTGNSCG
Mb3692c|M.bovis_AF2122/97 GLDYAVIKFDPAKVTPVAVFNGFAINGIGPDPSFGQIACKQGRTTGNSCG
Rv3668c|M.tuberculosis_H37Rv GLDYAVIKFDPAKVTPVAVFNGFAINGIGPDPSFGQIACKQGRTTGNSCG
MMAR_5156|M.marinum_M SLDYAVIKFDPAKVTPVANFNDFAINGIGPDPTFGQIACKQGRTTGNSCG
MUL_4242|M.ulcerans_Agy99 SLDYAVIKFDPAKVTPVANFNDFAINGIGPDPTFGQIACKQGRTTGNSCG
Mflv_1379|M.gilvum_PYR-GCK ILDYAVIKFDPAKVTPVNEVNGFRIDGLGPDPAFGEVACKLGRSTGYSCG
Mvan_5427|M.vanbaalenii_PYR-1 VLDYAVIKFDPARVTPVNTVNGFRIDGLGPDPRFGDVACKLGRSTGYSCG
TH_0965|M.thermoresistible__bu ALDYAVIEFDPQKVIPVNEVNGFRIDGLGPDPVFGEIACKLGRTTGHSCG
MSMEG_6180|M.smegmatis_MC2_155 LLDYAVIEFDPQKVTPTNNINGFRIDGIGPDPRFGEIACKLGRTTGYSCG
MAB_0424|M.abscessus_ATCC_1997 QYDYATIQFDPEKVQPVSTYKGFTITGIGPDPQPGDIACKLGRTTGNSCG
***.*:*** :* *. :.* * *:* ** *::*** **:** ***
MLBr_02295|M.leprae_Br4923 VAWGMGETSGTLVMQVCGRPGDSGAPVTVNNLLVGMIHGAFTDNLPVCIT
MAV_0461|M.avium_104 VTWGMGQSPGTIVMQVCGQPGDSGAPVTVNNQLVGMIHGAFSDNLPTCVI
Mb3692c|M.bovis_AF2122/97 VTWGPGESPGTLVMQVCGGPGDSGAPVTVDNLLVGMIHGAFSDNLPSCIT
Rv3668c|M.tuberculosis_H37Rv VTWGPGESPGTLVMQVCGGPGDSGAPVTVDNLLVGMIHGAFSDNLPSCIT
MMAR_5156|M.marinum_M VTWGPGQDPGTILMQVCGGPGDSGAPVTVNNMLVGMIHGAFSDSLPTCIT
MUL_4242|M.ulcerans_Agy99 VTWGPGQDPGTILMQVCGGPGDSGAPVTVNNMLVGMIHGAFSDSLPTCIT
Mflv_1379|M.gilvum_PYR-GCK VTWGPGQEPGTILNQVCGGPGDSGGPVTVNNQLVGMLHGAFSEDLPTCVV
Mvan_5427|M.vanbaalenii_PYR-1 VTWGPGQEPGTILNQVCGGPGDSGGPVTVNNRLVGMLHGAFSEDLPTCVV
TH_0965|M.thermoresistible__bu VTWGPGQQPGTIVNQVCGQPGDSGAPVTVNNRLVGMIHGAFSEVLPTCVV
MSMEG_6180|M.smegmatis_MC2_155 VTWGPGKDPGTIVNQVCGQPGDSGAPVTVNNLLVGMIHGAFTEDLPTCVV
MAB_0424|M.abscessus_ATCC_1997 VTFGAGAEPGTFINQVCGQPGDSGAPVTVNGQLVGMIHGAATK-LPVCVI
*::* * .**:: **** *****.****:. ****:*** :. ** *:
MLBr_02295|M.leprae_Br4923 KFIPLHTPAVVMSMNAILADVNKNNR-PGAGFVPQPV--
MAV_0461|M.avium_104 KYIPLHTPAVVMSFNAILADINAKHR-PGAGFAPLPG--
Mb3692c|M.bovis_AF2122/97 KYIPLHTPAVVMSINADLADINAKNR-PGAGFVPVPA--
Rv3668c|M.tuberculosis_H37Rv KYIPLHTPAVVMSINADLADINAKNR-PGAGFVPVPA--
MMAR_5156|M.marinum_M KYIPLHTPAVVMSMNADLDDIAAKNR-PGSGFVPVPG--
MUL_4242|M.ulcerans_Agy99 KYIPLHTPAVVMSMNADLDDIAAKNR-PGSGFVSVPG--
Mflv_1379|M.gilvum_PYR-GCK KFIPLHTPAVTMSINTQLADITAKGR-PGSGFVPVGAR-
Mvan_5427|M.vanbaalenii_PYR-1 KFIPLHTPAVTMSFNTQLADITAKGR-PGSGFVPVGATY
TH_0965|M.thermoresistible__bu KFIPLHTPAVTISFNTQLADINAKNR-PGAGFVPIGAVP
MSMEG_6180|M.smegmatis_MC2_155 KFVPLHTPAVTMSINTQLADITAKNR-PGTGFVPIR---
MAB_0424|M.abscessus_ATCC_1997 KYIPLHTPTTTYSINTVLADINAKNRTAGVGFTPVG---
*::*****:.. *:*: * *: : * .* **..