For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRDATRDELAADLAQAERSRDPIGQLTAAHPEIDVVDAYEIQLINIRQRVAEGARVVGHKVGLSSPIMQQ MMGVDEPDYGHLLDDMQVFEDTPVQASRYLSPRVEVEVGFILAADLPGAGCTEDDVLAATEALVPAIELI DTRIKDWQIKICDTIADNASAAGFVLGAARVPPADLDVRAIDAKLTRNGEVVAEGRSDAVLGNPATAVAW LAGKVESFGVRLRKGDIVLPGSCTFAVEARAGDEFVADFTGLGLVRLSFE*
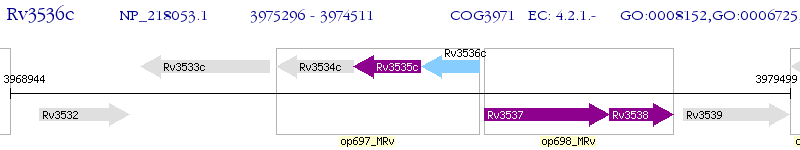
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3536c | - | - | 100% (261) | PROBABLE HYDRATASE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3566c | - | 1e-146 | 100.00% (261) | hydratase |
| M. gilvum PYR-GCK | Mflv_1520 | - | 1e-114 | 77.78% (261) | 4-oxalocrotonate decarboxylase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0624 | - | 1e-104 | 70.88% (261) | hydratase/decarboxylase |
| M. marinum M | MMAR_5023 | - | 1e-126 | 85.44% (261) | 2-keto-4-pentenoate hydratase |
| M. avium 104 | MAV_0625 | - | 1e-121 | 82.38% (261) | 2-keto-4-pentenoate hydratase |
| M. smegmatis MC2 155 | MSMEG_5940 | - | 1e-113 | 78.16% (261) | 2-keto-4-pentenoate hydratase |
| M. thermoresistible (build 8) | TH_0369 | - | 1e-113 | 78.16% (261) | PROBABLE HYDRATASE |
| M. ulcerans Agy99 | MUL_4097 | - | 1e-126 | 85.44% (261) | 2-keto-4-pentenoate hydratase |
| M. vanbaalenii PYR-1 | Mvan_5236 | - | 1e-111 | 77.39% (261) | 4-oxalocrotonate decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3536c|M.tuberculosis_H37Rv --------------------------------------------------
Mb3566c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_5023|M.marinum_M --------------------------------------------------
MUL_4097|M.ulcerans_Agy99 MRRQGQPPCRRRRFRPRRRRSPVMSCRTRYRIARVGRYWPGHHDARKTRG
MAV_0625|M.avium_104 --------------------------------------------------
MSMEG_5940|M.smegmatis_MC2_155 --------------------------------------------------
TH_0369|M.thermoresistible__bu --------------------------------------------------
Mflv_1520|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5236|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0624|M.abscessus_ATCC_1997 --------------------------------------------------
Rv3536c|M.tuberculosis_H37Rv ---MLRDATRDELAADLAQAERSRDPIGQLTAAHPEIDVVDAYEIQLINI
Mb3566c|M.bovis_AF2122/97 ---MLRDATRDELAADLAQAERSRDPIGQLTAAHPEIDVVDAYEIQLINI
MMAR_5023|M.marinum_M ---MLADSTRDKIAADLAQAERSRQPITPLTPSYPDIDVVDAYQIQLINI
MUL_4097|M.ulcerans_Agy99 LREMLADSTRDKIAADLAQAERSRQPITPLTPSYPDIDVVDAYQIQLINI
MAV_0625|M.avium_104 ---MLSVATRDELAAELAQAERSGEPIPPLTAAYPEIDVVDAYEIQLINI
MSMEG_5940|M.smegmatis_MC2_155 ---MLSDEVRARLAADLAQAERSRQPITPLTDGNPDIDVVDAYEIQLINI
TH_0369|M.thermoresistible__bu ---MLSAQVREQLAAELAQAERSRVPITPLTDRHPGIDVVDAYEIQLINI
Mflv_1520|M.gilvum_PYR-GCK ---MLPAEVREKLAADLAEAEQSRKPISPLTDAHPDIDVVDAYEIQLINI
Mvan_5236|M.vanbaalenii_PYR-1 ---MLSVEVREKLAADLAEAERSRVPISPLTDGQPGIDVVDAYEIQLINI
MAB_0624|M.abscessus_ATCC_1997 ---MLSSEVRDSIAADLAEAERTRVAIAPPTDTHPDFDVVDAYEIQLMNI
** .* :**:**:**:: .* * * :******:***:**
Rv3536c|M.tuberculosis_H37Rv RQRVAEGARVVGHKVGLSSPIMQQMMGVDEPDYGHLLDDMQVFEDTPVQA
Mb3566c|M.bovis_AF2122/97 RQRVAEGARVVGHKVGLSSPIMQQMMGVDEPDYGHLLDDMQVFEDTPVQA
MMAR_5023|M.marinum_M RRRVAEGGRVVGHKVGLSSPVMQQMMGVDEPDYGHLLDNMRVFEDAPVRT
MUL_4097|M.ulcerans_Agy99 RRRVAEGGRVVGHKVGLSSPVMQQMMGVDEPDYGHLLDNMRVFEDAPVRT
MAV_0625|M.avium_104 RQRVAEGARVLGHKVGLSSLAIQQMMGVDEPDYGHLLDDMQLFEDTPVKT
MSMEG_5940|M.smegmatis_MC2_155 RQRIAEGAKVVGHKVGLSSKAMQEMMGVDEPDYGHLLDDMQVYEDQPVKS
TH_0369|M.thermoresistible__bu RQRVAEGARVVGHKVGLSSEAMQKMMGVDEPDYGHLLDDMQVFEDKPVKS
Mflv_1520|M.gilvum_PYR-GCK RRRVADGARIIGHKVGLSSEAMQKMMGVDEPDYGHLLEEMQVFEDVPVGS
Mvan_5236|M.vanbaalenii_PYR-1 RQRVAEGARVIGHKVGLSSEAMQKMMNVDEPDYGHLLDEMQVFENVAVKS
MAB_0624|M.abscessus_ATCC_1997 RSRLADGAKVVGHKVGLSSEAMQKMMGVHEPDYGHLLADMELFEDVAASA
* *:*:*.:::******** :*:**.*.******** :*.::*: .. :
Rv3536c|M.tuberculosis_H37Rv SRYLSPRVEVEVGFILAADLPGAGCTEDDVLAATEALVPAIELIDTRIKD
Mb3566c|M.bovis_AF2122/97 SRYLSPRVEVEVGFILAADLPGAGCTEDDVLAATEALVPAIELIDTRIKD
MMAR_5023|M.marinum_M ANYLYPRVEVEVGFILAADLPGADCTEDDVLAATEALVPSIELIDTRIKD
MUL_4097|M.ulcerans_Agy99 ANYLYPRVEVEVGFILAADLPGADCTEDDVLAATEALVPSIELIDTRIKD
MAV_0625|M.avium_104 NRYLYPRVEVEVGFILNADLPGAGCTEDDVMAATEAFVPAIELIDTRITD
MSMEG_5940|M.smegmatis_MC2_155 ANYLYPRVEVEVGFILADDLPGAGCTEEDVLAATAAFAPAIELIDTRITN
TH_0369|M.thermoresistible__bu ANYLYPRVEVEVGFILAADLPGADCTEEDVLAATEAFAPAIELIDTRITD
Mflv_1520|M.gilvum_PYR-GCK GRYLYPRVEVEVGFILSDDLPGAGCTEDDVLAATAAFAPAIELIDTRITN
Mvan_5236|M.vanbaalenii_PYR-1 ANYLYPRVEVEVGFILSDDLPGAGCTEDDVLAATAAFAPAIELIDTRITN
MAB_0624|M.abscessus_ATCC_1997 SKFLFPRVEVEVGFILAADLPGEDCTEDDVLAATAAYVPSIEVVDSRITN
.:* *********** **** .***:**:*** * .*:**::*:**.:
Rv3536c|M.tuberculosis_H37Rv WQIKICDTIADNASAAGFVLGAARVPPADLDVRAIDAKLTRNGEVVAEGR
Mb3566c|M.bovis_AF2122/97 WQIKICDTIADNASAAGFVLGAARVPPADLDVRAIDAKLTRNGEVVAEGR
MMAR_5023|M.marinum_M WQIKICDTIADNASAAGFVLGAARVSPAEVDVKAIDAVLTRNGELAAEGR
MUL_4097|M.ulcerans_Agy99 WQIKICDTIADNASAAGFVLGAARVSPAEVDVKAIDAVLTRNGELAAEGR
MAV_0625|M.avium_104 WKIELCDTIADNASSAGFVLGAARVSPQDIDIKGIDAVLRRNGEVVAEGR
MSMEG_5940|M.smegmatis_MC2_155 WQIKLCDTIADNASSAGWVLGEARVSPKDIDIRNIDAVLKNNGEVVAEGR
TH_0369|M.thermoresistible__bu WKIKLCDTIADNASSAGWVLGEARVSPEEIDITTIEATLTNNGEVVAQGR
Mflv_1520|M.gilvum_PYR-GCK WQIKLCDTIADNASSAGWVLGEARVSPKDVDIRAIDAVLTNNGEVVAEGR
Mvan_5236|M.vanbaalenii_PYR-1 WQIKLCDTIADNASSAGWVLGEARVSPKDVDIKNIDAVLTNNGEVVAEGR
MAB_0624|M.abscessus_ATCC_1997 WQIKICDTIADNASSAGFVVGKQRVSPKDIDIKSIDAVLTCNGEKLAEGR
*:*::*********:**:*:* **.* ::*: *:* * *** *:**
Rv3536c|M.tuberculosis_H37Rv SDAVLGNPATAVAWLAGKVESFGVRLRKGDIVLPGSCTFAVEARAGDEFV
Mb3566c|M.bovis_AF2122/97 SDAVLGNPATAVAWLAGKVESFGVRLRKGDIVLPGSCTFAVEARAGDEFV
MMAR_5023|M.marinum_M SDAVLGNPATAVAWLARKVEGFGVRLRKGDIVLPGSCTFAVDANAGDEFV
MUL_4097|M.ulcerans_Agy99 SDAVLGNPATAVAWLARKVEGFGVRLRKGDIVLPGSCTFAVDANAGDEFV
MAV_0625|M.avium_104 TDAVLGNPVTAVAWLARKVDGFGVRLRKGDVVLPGSCTKAIDAHPGDEFV
MSMEG_5940|M.smegmatis_MC2_155 SDAVLGNPVTAVAWLARKVESFGVRLKAGDIVLPGSCTRAIDAPAGSDFV
TH_0369|M.thermoresistible__bu SDAVLGNPVTAVAWLARKVDSFGVRLKAGDIVLPGSCTRAIDAPAGSHFV
Mflv_1520|M.gilvum_PYR-GCK SDAVLGNPVTAVAWLARKVEAFGVRLKAGDIVLPGSCTRAIDAPPGSRFV
Mvan_5236|M.vanbaalenii_PYR-1 SDAVLGNPVTAVAWLARKVESFGVRLKAGDIVLPGSCTRAIDAPAGSHFV
MAB_0624|M.abscessus_ATCC_1997 SDAVLGNPVTSVAWLARKVASFGVRLRAGDVVLPGSCTRAFDAHPGDEFL
:*******.*:***** ** .*****: **:******* *.:* .*. *:
Rv3536c|M.tuberculosis_H37Rv ADFTGLGLVRLSFE
Mb3566c|M.bovis_AF2122/97 ADFTGLGLVRLSFE
MMAR_5023|M.marinum_M ADFTGLGSVRLSFQ
MUL_4097|M.ulcerans_Agy99 ADFTGLGSVRLSFQ
MAV_0625|M.avium_104 ADFAGLGSVRLSFE
MSMEG_5940|M.smegmatis_MC2_155 ADFAGLGSVHLRFE
TH_0369|M.thermoresistible__bu ADFSGLGSVQLSFE
Mflv_1520|M.gilvum_PYR-GCK ADFTGLGSVRLDFE
Mvan_5236|M.vanbaalenii_PYR-1 ADFTGLGSVRLDFE
MAB_0624|M.abscessus_ATCC_1997 AEFAGLGSVRLAFE
*:*:*** *:* *: