For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRISYTPQQEELRRELRSYFATLMTPERREALSSVQGEYGVGNVYRETIAQMGRDGWLALGWPKEYGGQG RSAMDQLIFTDEAAIAGAPVPFLTINSVAPTIMAYGTDEQKRFFLPRIAAGDLHFSIGYSEPGAGTDLAN LRTTAVRDGDDYVVNGQKMWTSLIQYADYVWLAVRTNPESSGAKKHRGISVLIVPTTAEGFSWTPVHTMA GPDTSATYYSDVRVPVANRVGEENAGWKLVTNQLNHERVALVSPAPIFGCLREVREWAQNTKDAGGTRLI DSEWVQLNLARVHAKAEVLKLINWELASSQSGPKDAGPSPADASAAKVFGTELATEAYRLLMEVLGTAAT LRQNSPGALLRGRVERMHRACLILTFGGGTNEVQRDIIGMVALGLPRANR
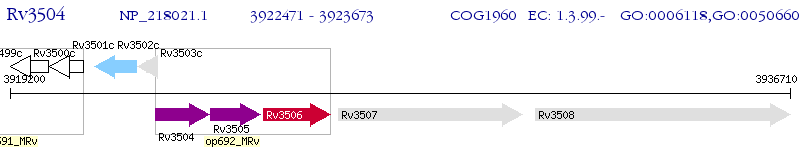
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3504 | fadE26 | - | 100% (400) | PROBABLE ACYL-CoA DEHYDROGENASE FADE26 |
| M. tuberculosis H37Rv | Rv3543c | fadE29 | 9e-91 | 45.52% (402) | acyl-CoA dehydrogenase FADE29 |
| M. tuberculosis H37Rv | Rv1934c | fadE17 | 6e-46 | 32.55% (424) | acyl-CoA dehydrogenase FADE17 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3534 | fadE26 | 0.0 | 100.00% (400) | acyl-CoA dehydrogenase FADE26 |
| M. gilvum PYR-GCK | Mflv_1551 | - | 0.0 | 82.50% (400) | acyl-CoA dehydrogenase domain-containing protein |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-28 | 28.45% (355) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4158 | - | 0.0 | 84.25% (400) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4992 | fadE26 | 0.0 | 90.00% (400) | acyl-CoA dehydrogenase FadE26 |
| M. avium 104 | MAV_0652 | - | 0.0 | 87.91% (397) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5906 | - | 0.0 | 83.25% (400) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3993 | - | 0.0 | 85.11% (403) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4067 | fadE26 | 0.0 | 89.50% (400) | acyl-CoA dehydrogenase FadE26 |
| M. vanbaalenii PYR-1 | Mvan_5206 | - | 0.0 | 84.25% (400) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3504|M.tuberculosis_H37Rv MRISYTP----QQEELRRELRSYFATLMTPERREALSSVQGEYGVGNVYR
Mb3534|M.bovis_AF2122/97 MRISYTP----QQEELRRELRSYFATLMTPERREALSSVQGEYGVGNVYR
MMAR_4992|M.marinum_M MRISYTP----EQEELRRELRSYFTTLMTPERREALSSVQGEYGTGNVYR
MUL_4067|M.ulcerans_Agy99 MRISYTP----EQEELRRELRSYFTTLMTPERREALSSVQGEYGTGNVYR
MAV_0652|M.avium_104 MRISYTP----EQEDLRRELRSYFTKLMTPERREALSSTQGEYGVGNVYR
Mflv_1551|M.gilvum_PYR-GCK MRIGYTP----EQEELRRELRSYFTKLMTPERAEALAASDGEMGRGNVYR
Mvan_5206|M.vanbaalenii_PYR-1 MRIGYTP----EQEELRRELRAYFTKLMTPERAEALAASDGEMGRGNVYR
MSMEG_5906|M.smegmatis_MC2_155 MRIGYTP----EQEELRRELRAYFSKLMTPERVEALSSSEGEMGRGNVYR
TH_3993|M.thermoresistible__bu MRIGYTP----EQEELRRELRAYFSKLMTPDRVEALNSAEGEYGRGDVYR
MAB_4158|M.abscessus_ATCC_1997 MHIAYTP----EQEELRRELRAYFDKLLTPERREALASNQGEYGSGNVYR
MLBr_00737|M.leprae_Br4923 -MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPE
:.:: : .* .* . .: : . * . . . .
Rv3504|M.tuberculosis_H37Rv ETIAQMGRDGWLALGWPKEYGGQGRSAMDQLIFTDEAAIAGAPVPFLTIN
Mb3534|M.bovis_AF2122/97 ETIAQMGRDGWLALGWPKEYGGQGRSAMDQLIFTDEAAIAGAPVPFLTIN
MMAR_4992|M.marinum_M ETVAQMGKDGWLTLNWPKEYGGQDRSPMDSLIFTDEAAIAGAPVPFLTIN
MUL_4067|M.ulcerans_Agy99 ETVAQMGKNGWLTLNWPKEYGGQDRSPMDSLIFTDEAAIAGAPVPFLTIN
MAV_0652|M.avium_104 ETVAQMGKDGWLTLNWPKEYGGQDRSPMDSLIFTDEAAIAGAPVPFLTIN
Mflv_1551|M.gilvum_PYR-GCK ETVAQMGKDGWLTLSWPEEFGGQARPPMDGLIFNDEAAIANVPVPFLTIN
Mvan_5206|M.vanbaalenii_PYR-1 ETVAQMGKDGWLTLSWPKEFGGQARPPMDGLIFNDEAAIANVPVPFLTIN
MSMEG_5906|M.smegmatis_MC2_155 ETVAQMGKDGWLTLSWPKEFGGQARPPMDGLIFTDEAAIAGAPVPFLTIN
TH_3993|M.thermoresistible__bu ETVAQMGRDGWLALGWPKEYGGQARSAMDQLIFTDEAAIAGAPVPFLTIN
MAB_4158|M.abscessus_ATCC_1997 ETVEQMGADGWLALGWPKEFGGQDRSVMDQLIFTDEAAIAGAPVPFLTIN
MLBr_00737|M.leprae_Br4923 EALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAV
*:: :. .*: :: *:*:*** : *. :*.* ......::.
Rv3504|M.tuberculosis_H37Rv SVAPTI--MAYGTDEQKRFFLPRIAAGDLHFSIGYSEPGAGTDLANLRTT
Mb3534|M.bovis_AF2122/97 SVAPTI--MAYGTDEQKRFFLPRIAAGDLHFSIGYSEPGAGTDLANLRTT
MMAR_4992|M.marinum_M SVAPTI--MAFGTDEQKKFFLPKIAAGDLHFSIGYSEPGAGTDLANLRTT
MUL_4067|M.ulcerans_Agy99 SVAPTI--MAFGTDEQKKFFLPKIAAGDLHFSIGYSEPGAGTDLANLRTT
MAV_0652|M.avium_104 SVAPTI--MAFGTDEQKKYFLPKIAAGELHFSIGYSEPGAGTDLANLRTT
Mflv_1551|M.gilvum_PYR-GCK SVAPTI--MHFGTEEQKKFFLPRIAKGDLHFSIGYSEPGAGTDLASLRTT
Mvan_5206|M.vanbaalenii_PYR-1 SVAPTI--MAFGTEEQKKFFLPKIAAGDLHFSIGYSEPGAGTDLAALRTT
MSMEG_5906|M.smegmatis_MC2_155 SVAPTI--MAFGTEEQKKFFLPKIASGELHFSIGYSEPGAGTDLASLRTT
TH_3993|M.thermoresistible__bu SVAPTI--MHFGTEEQKKFFLPKIAAGELHFSIGYSEPGAGTDLASLRTT
MAB_4158|M.abscessus_ATCC_1997 SVAPTI--MHFGTDEQKKFFLPKIAAGKLHFSIGYSEPGAGTDLASLRTS
MLBr_00737|M.leprae_Br4923 NKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTR
. *: : *::* *: .** :* * . ** **:* * :**
Rv3504|M.tuberculosis_H37Rv AVRDGDDYVVNGQKMWTSLIQYADYVWLAVRTNPESSGAKKHRGISVLIV
Mb3534|M.bovis_AF2122/97 AVRDGDDYVVNGQKMWTSLIQYADYVWLAVRTNPESSGAKKHRGISVLIV
MMAR_4992|M.marinum_M AVRDGDDYVINGQKMWTSLIAYADWVWLAVRTNPE---AKKHRGISMLVV
MUL_4067|M.ulcerans_Agy99 AVRDGDDYLINGQKMWTSLIAYADWVCLAVRTNPE---AKKHRGISMLVV
MAV_0652|M.avium_104 AVRDGDDYVINGQKMWTSLIAYADWVWLAVRTNPE---AKKHRGISMLVV
Mflv_1551|M.gilvum_PYR-GCK AVHDGDDYVINGQKMWTSLIAYADYVWLAVRTNTE---AKKHRGISMLIV
Mvan_5206|M.vanbaalenii_PYR-1 AVRDGDDYVINGQKMWTSLIAYADYVWLAVRTNPE---AKKHRGISMLIV
MSMEG_5906|M.smegmatis_MC2_155 AVRDGDDYVINGQKMWTSLIAYADYVWLAVRTNPE---AKKHRGISMLIV
TH_3993|M.thermoresistible__bu AVRDGDEYVINGQKMWTSLIQYADYVWLAVRTNPE---AKKHRGISMLIV
MAB_4158|M.abscessus_ATCC_1997 AVRDGDDYIVNGQKMWTSLIEYADYIWLAVRTNTE---VKKHRGISMLIV
MLBr_00737|M.leprae_Br4923 AKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPD----KGANGISAFIV
* ***::::** * * : : : : . *:.: * .*** ::*
Rv3504|M.tuberculosis_H37Rv PTTAEGFSWTPVHT---MAGPDTSATYYSDVRVPVANRVGEENAGWKLVT
Mb3534|M.bovis_AF2122/97 PTTAEGFSWTPVHT---MAGPDTSATYYSDVRVPVANRVGEENAGWKLVT
MMAR_4992|M.marinum_M PTTAEGFSWTPVHT---MAGPDTSATYYTDVRVPVTNLIGEENAGWKLVT
MUL_4067|M.ulcerans_Agy99 PTTAEGFSWTPVHT---MAGPDTSATYYTDVRVPVTNLIGEENAGWKLVT
MAV_0652|M.avium_104 PTTAEGFSWTPVHT---MAGPDTSATYYSDVRVPVTNLVGEENGGWKLVT
Mflv_1551|M.gilvum_PYR-GCK PTTAEGFSWTPVHT---VSGVDTSATYYQDVRVPKSALVGEENGGWKLVT
Mvan_5206|M.vanbaalenii_PYR-1 PTTAEGFSWTPVHT---MSGVDTSATYYQDVRVPVSSLVGEENAGWKLVT
MSMEG_5906|M.smegmatis_MC2_155 PTTAEGFSWTPVHT---MSGVDTSATYYQDVRVPVTNLVGEENQGWKLVT
TH_3993|M.thermoresistible__bu PTTAEGFSWTPVHT---MAGVDTSATYYQDVRVPVSNRVGEENEGWKLVT
MAB_4158|M.abscessus_ATCC_1997 PTTAEGFSYTKVHT---MAGPGTSATYYQDVRVPVTSRVGEENAGWKLVT
MLBr_00737|M.leprae_Br4923 HKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTAL
. **** .. : * *: *: . *:* :** . *:* .
Rv3504|M.tuberculosis_H37Rv NQLNHERVALVSPAP--IFGCLREVREWAQN---TKDAGGTRLIDSEWVQ
Mb3534|M.bovis_AF2122/97 NQLNHERVALVSPAP--IFGCLREVREWAQN---TKDAGGTRLIDSEWVQ
MMAR_4992|M.marinum_M NQLNHERVALVSSAP--ISMALREVREWAQN---TKDAGGARLIDSEWVQ
MUL_4067|M.ulcerans_Agy99 NQLNHERVALVSSAP--ISMALREVREWAQN---TKDAGGARLIDSEWVQ
MAV_0652|M.avium_104 NQLNHERVALVSPAP--IYLALREVREWAQN---TKDASGARLIDSEWVQ
Mflv_1551|M.gilvum_PYR-GCK NQLNHERVALVSAQP--IFVALNGVREWAQN---TKDVHGNRVIDSEWVQ
Mvan_5206|M.vanbaalenii_PYR-1 NQLNHERVALVSAQP--IFVALNGVREWAQN---TKDVHGKRLIDSEWVQ
MSMEG_5906|M.smegmatis_MC2_155 NQLNHERVALVSAQP--IYVALNGVREWAQN---TKDVHGKRLIDSEWVQ
TH_3993|M.thermoresistible__bu NQLNHERVALVSAQP--IFLALDGVREWAQSTHISPDGRGPRVIDAEWVQ
MAB_4158|M.abscessus_ATCC_1997 NQLNHERVALVSSAP--IITALREVREWAQN---TKGPGG-RIIDAEWVQ
MLBr_00737|M.leprae_Br4923 ATLDHTRPTIGAQAVGIAQGALDAAIVYTKD----RKQFGESISTFQSIQ
*:* * :: : .* . :::. * : : :*
Rv3504|M.tuberculosis_H37Rv LNLARVHAKAEVLKLINWELASSQSGPKDAGPSPADASAAKVFGTELATE
Mb3534|M.bovis_AF2122/97 LNLARVHAKAEVLKLINWELASSQSGPKDAGPSPADASAAKVFGTELATE
MMAR_4992|M.marinum_M LNLARVHAKVEVLKLINWELASND-----AAPSPADASAAKVFGTELATE
MUL_4067|M.ulcerans_Agy99 LNLARVHAKVEVLKLINWELASND-----AAPSPADASAAKVFGTELATE
MAV_0652|M.avium_104 INLARVHAKAEVLKLINWELASSDG----HALGPADASAAKVYGTELATE
Mflv_1551|M.gilvum_PYR-GCK LNLARVHAKAEVLKLINWELASSTD----TAPSPADASAAKVFGTELATE
Mvan_5206|M.vanbaalenii_PYR-1 LNLARVHAKAEVLKLINWELASATD----AAPSPADASAAKVYGTELATE
MSMEG_5906|M.smegmatis_MC2_155 LNLARVHAKAEVLKLINWELAS-TE----AAPSPADASAAKVYGTELATE
TH_3993|M.thermoresistible__bu LNLARVHAKAEVLKLINWELASSDA----AAPSPADASAAKVFGTELATE
MAB_4158|M.abscessus_ATCC_1997 LNLARVHAKAEYLKLINWELASTTD----AAPSPADASATKVFGTELATE
MLBr_00737|M.leprae_Br4923 FMLADMAMKVEAARLIVYAAAARAE--RGEPDLGFISAASKCFASDIAME
: ** : *.* :** : *: ::*:* :.:::* *
Rv3504|M.tuberculosis_H37Rv AYRLLMEVLGTAATLRQNSPGALLRGRVERMHRACLILTFGGGTNEVQRD
Mb3534|M.bovis_AF2122/97 AYRLLMEVLGTAATLRQNSPGALLRGRVERMHRACLILTFGGGTNEVQRD
MMAR_4992|M.marinum_M AYRLLMEVLGTAATLRQDSPGALLHGRVERMHRACLILTFGGGTNEVQRD
MUL_4067|M.ulcerans_Agy99 AYRLLMEVLGTAATLRQDSPGALLRGRVERMHRACLILTFGGGTNEVQRD
MAV_0652|M.avium_104 AYRLLMEVLGTGATVRQDSPGALLRGRVERMHRACLILTFGGGTNEVQRD
Mflv_1551|M.gilvum_PYR-GCK AYRLLMEVLGTAATLRTNSPGAVLRGRIERMHRSCLILTFGGGTNEIQRD
Mvan_5206|M.vanbaalenii_PYR-1 AYRLLMEVLGTAATLRTNSPGALLRGRIERMHRSCLILTFGGGTNEIQRD
MSMEG_5906|M.smegmatis_MC2_155 AYRLLMEVLGTSATLRGDTRGALLRGRVERFHRSALILTFGGGTNEIQRD
TH_3993|M.thermoresistible__bu AYRLLMEVLGTRATLRPDSPGALLRGRIERMHRSCLILTFGGGTNEVQRD
MAB_4158|M.abscessus_ATCC_1997 AYRLLMEVLGTAATLRQDSPGAQLRGRVERMHRACLILTFGGGTNEVQRD
MLBr_00737|M.leprae_Br4923 VTTDAVQLFG-GAGYTSDFP-------VERFMRDAKITQIYEGTNQIQRV
. ::::* * : :**: * . * : ***::**
Rv3504|M.tuberculosis_H37Rv IIGMVALGLPRANR
Mb3534|M.bovis_AF2122/97 IIGMVALGLPRANR
MMAR_4992|M.marinum_M IIGMVALGLPRANR
MUL_4067|M.ulcerans_Agy99 IIGMVALGLPRANR
MAV_0652|M.avium_104 IIGMVALGLPR-NR
Mflv_1551|M.gilvum_PYR-GCK IIGMVALGLPRVNR
Mvan_5206|M.vanbaalenii_PYR-1 IIGMVALGLPRVNR
MSMEG_5906|M.smegmatis_MC2_155 IIGMVALGLPRVNR
TH_3993|M.thermoresistible__bu IIGMVALGLPRVNR
MAB_4158|M.abscessus_ATCC_1997 IIAMTALGQPAASR
MLBr_00737|M.leprae_Br4923 VMSRALLR------
::. . *