For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNAPKIKVSGPVVELDGDEMTRVIWKLIKDMLILPYLDIRLDYYDLGIEHRDATDDQVTIDAAYAIKKHG VGVKCATITPDEARVEEFNLKKMWLSPNGTIRNILGGTIFREPIVISNVPRLVPGWTKPIVIGRHAFGDQ YRATNFKVDQPGTVTLTFTPADGSAPIVHEMVSIPEDGGVVLGMYNFKESIRDFARASFSYGLNAKWPVY LSTKNTILKAYDGMFKDEFERVYEEEFKAQFEAAGLTYEHRLIDDMVAACLKWEGGYVWACKNYDGDVQS DTVAQGYGSLGLMTSVLMTADGKTVEAEAAHGTVTRHYRQYQAGKPTSTNPIASIFAWTRGLQHRGKLDG TPEVIDFAHKLESVVIATVESGKMTKDLAILIGPEQDWLNSEEFLDAIADNLEKELAN*
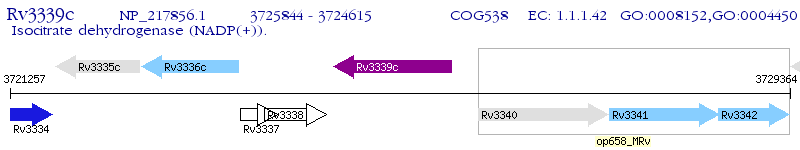
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3339c | icd1 | - | 100% (409) | isocitrate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3371c | icd1 | 0.0 | 100.00% (409) | isocitrate dehydrogenase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1186 | icd1 | 0.0 | 90.66% (407) | isocitrate dehydrogenase |
| M. avium 104 | MAV_4311 | - | 0.0 | 88.48% (408) | isocitrate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3285 | - | 3e-05 | 24.88% (213) | 3-isopropylmalate dehydrogenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3339c|M.tuberculosis_H37Rv MS---NAPKIKVSGPVVELDGDEMTRVIWKLIKDMLILPYLDIRLDYYDL
Mb3371c|M.bovis_AF2122/97 MS---NAPKIKVSGPVVELDGDEMTRVIWKLIKDMLILPYLDIRLDYYDL
MAV_4311|M.avium_104 MSKPANTSKIKVKGRVVELDGDEMTRVIWKMIKENLILPYLDIDLDYYDL
MMAR_1186|M.marinum_M MS---GEPKITVSGPVVELDGDEMTRVIWKLIKDRLILPHLDVKLLYYDL
MSMEG_3285|M.smegmatis_MC2_155 --------MIELGVLRGDGIGPEIVSATTQVVERALDAAAVGVRWVPLPF
* : : * *:. . :::: * . :.: :
Rv3339c|M.tuberculosis_H37Rv G---IEHRDATDDQVTIDAAYAIKKHGVGVKCATITPDEARVEEFNLKKM
Mb3371c|M.bovis_AF2122/97 G---IEHRDATDDQVTIDAAYAIKKHGVGVKCATITPDEARVEEFNLKKM
MAV_4311|M.avium_104 G---IEHRDRTDDQVTIDAAYAIKKHGVGVKCATITPDEARVREFNLKRM
MMAR_1186|M.marinum_M G---IEHRDRTNDQVTIDAAYAIKKHGVGVKCATITPDEARVREFNLKKM
MSMEG_3285|M.smegmatis_MC2_155 GRQAIDEFGTPVPDVTLERLDGLTGWIIGPHDNAGYPEQFRG--------
* *:. . . :**:: .:. :* : : *:: *
Rv3339c|M.tuberculosis_H37Rv WLSPNGTIRN--ILGGTIFREPIVISNVPRLVPGWTKPIVIGRHAFGDQY
Mb3371c|M.bovis_AF2122/97 WLSPNGTIRN--ILGGTIFREPIVISNVPRLVPGWTKPIVIGRHAFGDQY
MAV_4311|M.avium_104 WLSPNGTIRN--ILGGTIFREPIVISNVPRLVPGWSKPIVIGRHAFGDQY
MMAR_1186|M.marinum_M WLSPNGTIRN--ILGGTIFREPIVISNVPRLVPGWTKPIVIGRHAFGDQY
MSMEG_3285|M.smegmatis_MC2_155 TLSPGGVIRKRYNLFANIRPARALSKSVRAVCP--DIDVVIVRENTEGFY
***.*.**: * ..* : ..* : * :** *. . *
Rv3339c|M.tuberculosis_H37Rv RATNFKVDQPGTVTLTFTPADGSAPIVHEMVSIPEDGGVVLGMYNFKESI
Mb3371c|M.bovis_AF2122/97 RATNFKVDQPGTVTLTFTPADGSAPIVHEMVSIPEDGGVVLGMYNFKESI
MAV_4311|M.avium_104 RSTNFKVDKPGTVAITFTPADGSEPIVHEMVSIPEDGGVVMGMYNFRESI
MMAR_1186|M.marinum_M RATNFKVDRPGTVTLTFTPADGSEPIVHEMVSIPEDGGVVMGMYNFRKSI
MSMEG_3285|M.smegmatis_MC2_155 ADRNMYDGAG-----EFMPTADVALAVAVFT---------------RAAI
*: . * *: . * :. : :*
Rv3339c|M.tuberculosis_H37Rv RDFARASFSYGLNAKWPVYLSTKNTILKAYDGMFKDEFERVYEEEFKAQF
Mb3371c|M.bovis_AF2122/97 RDFARASFSYGLNAKWPVYLSTKNTILKAYDGMFKDEFERVYEEEFKAQF
MAV_4311|M.avium_104 RDFARASFKYGLNAKWPVYLSTKNTILKAYDGMFKDEFQRIYDEEFKEQF
MMAR_1186|M.marinum_M QDFARASFSYGLNAKWPVYLSTKNTILKAYDGMFKDEFQRIYEEEFKERF
MSMEG_3285|M.smegmatis_MC2_155 ERIAHEAFRIAATRRRSVTIAHKTNVLTRTTGLFRDTCMAVADQYPDVEV
. :*: :* . . : .* :: *..:*. *:*:* : :: . ..
Rv3339c|M.tuberculosis_H37Rv EAAGLTYEHRLIDDMVAACLKWEGGYVWACKNYDGDVQSDTVAQGYGSLG
Mb3371c|M.bovis_AF2122/97 EAAGLTYEHRLIDDMVAACLKWEGGYVWACKNYDGDVQSDTVAQGYGSLG
MAV_4311|M.avium_104 EAEGLTYEHRLIDDMVAACLKWEGGYVWACKNYDGDVQSDAIAQGYGSLG
MMAR_1186|M.marinum_M DAEGLTYEHRLIDDMVAACLKWEGGYVWACKNYDGDVQSDTVAQGYGSLG
MSMEG_3285|M.smegmatis_MC2_155 RGE---HVDALAALLVSRPTQFD---VIVAENMFGDILSDLTGQLSGSLG
. : . * :*: ::: * ..:* **: ** .* ****
Rv3339c|M.tuberculosis_H37Rv LMTSVLMTADGKTVEAEAAHGTVTRHYRQYQAGKPTSTNPIASIFAWTRG
Mb3371c|M.bovis_AF2122/97 LMTSVLMTADGKTVEAEAAHGTVTRHYRQYQAGKPTSTNPIASIFAWTRG
MAV_4311|M.avium_104 LMTSVLMTADGKTVEAEAAHGTVTRHFRQYQAGKPTSTNPIASIFAWTRG
MMAR_1186|M.marinum_M LMTSVLMTADGKTVEAEAAHGTVTRHYRQYQAGQPTSTNPIASIFAWTRG
MSMEG_3285|M.smegmatis_MC2_155 MAPSLNASDS--KAMAQASHGAAP------DIAGRDVANPVAMMLSAAML
: .*: : . .. *:*:**:.. : . :**:* ::: :
Rv3339c|M.tuberculosis_H37Rv LQHRGKLDGTP-EVIDFAHKLESVVIATVESGKMTKDLAILIGPEQDWLN
Mb3371c|M.bovis_AF2122/97 LQHRGKLDGTP-EVIDFAHKLESVVIATVESGKMTKDLAILIGPEQDWLN
MAV_4311|M.avium_104 LAHRGKLDDTP-EVIEFAQTLERVVVSTVESGKMTKDLAILIGPDQKWQQ
MMAR_1186|M.marinum_M LAHRGKLDNTP-EVVQFAQTLEDVVIKTVESGKMTKDLAILIGPQQAWLN
MSMEG_3285|M.smegmatis_MC2_155 LRWLGERAGSGRALVGAADAVEHAVRRTLDAGLSTPDVGGSAG-------
* *: .: :: *. :* .* *:::* * *:. *
Rv3339c|M.tuberculosis_H37Rv SEEFLDAIADNLEKELAN
Mb3371c|M.bovis_AF2122/97 SEEFLDAIADNLEKELAN
MAV_4311|M.avium_104 SEEFLNSIAENLRRELTN
MMAR_1186|M.marinum_M SEEFLEAIAENLAKKLG-
MSMEG_3285|M.smegmatis_MC2_155 TREFTDGVLTALRPT---
:.** :.: *