For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNPIQAPDGGQGWPAAALGPAQRAVAWKRLGTEQFDVVVIGGGVVGSGCALDAATRGLKVALVEARDLAS GTSSRSSKMFHGGLRYLEQLEFGLVREALYERELSLTTLAPHLVKPLPFLFPLTKRWWERPYIAAGIFLY DRLGGAKSVPAQRHFTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTAAHYGAVVRCSTQVV ALLREGDRVIGVGVRDSENGAVAEVRGHVVVNATGVWTDEIQALSKQRGRFQVRASKGVHVVVPRDRIVS DVAMILRTEKSVMFVIPWGSHWIIGTTDTDWNLDLAHPAATKADIDYILGTVNAVLATPLTHADIDGVYA GLRPLLAGESDDTSKLSREHAVAVPAAGLVAIAGGKYTTYRVMAADAIDAAVQFIPARVAPSITEKVSLL GADGYFALVNQAEHVGALQGLHPYRVRHLLDRYGSLISDVLAMAASDPSLLSPITEAPGYLKVEAAYAAA AEGALHLEDILARRMRISIEYPHRGVDCAREVAEVVAPVLGWTAADIDREVANYMARVEAEVLSQAQPDD VSADMLRASAPEARAEILEPVPLD*
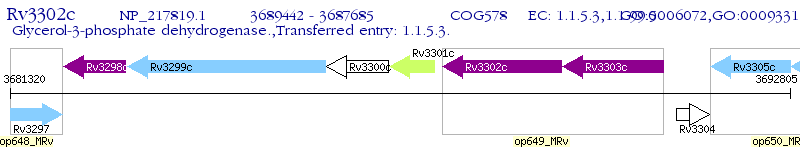
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3302c | glpD2 | - | 100% (585) | PROBABLE GLYCEROL-3-PHOSPHATE DEHYDROGENASE GLPD2 |
| M. tuberculosis H37Rv | Rv2249c | glpD1 | 2e-64 | 36.19% (514) | glycerol-3-phosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3330c | glpD2 | 0.0 | 100.00% (585) | glycerol-3-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4821 | - | 0.0 | 85.30% (585) | FAD dependent oxidoreductase |
| M. leprae Br4923 | MLBr_00713 | glpD | 0.0 | 90.92% (584) | putative glycerol-3-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3655c | - | 0.0 | 82.81% (576) | glycerol-3-phosphate dehydrogenase |
| M. marinum M | MMAR_1225 | glpD2 | 0.0 | 91.45% (585) | glycerol-3-phosphate dehydrogenase GlpD2 |
| M. avium 104 | MAV_4278 | - | 0.0 | 89.40% (585) | glycerol-3-phosphate dehydrogenase 2 |
| M. smegmatis MC2 155 | MSMEG_1736 | - | 0.0 | 85.47% (585) | glycerol-3-phosphate dehydrogenase 2 |
| M. thermoresistible (build 8) | TH_1838 | glpD2 | 0.0 | 84.38% (589) | PROBABLE GLYCEROL-3-PHOSPHATE DEHYDROGENASE GLPD2 |
| M. ulcerans Agy99 | MUL_2667 | glpD2 | 0.0 | 91.45% (585) | glycerol-3-phosphate dehydrogenase GlpD2 |
| M. vanbaalenii PYR-1 | Mvan_1620 | - | 0.0 | 85.37% (588) | FAD dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4821|M.gilvum_PYR-GCK MSDPIPAPGNGETF----LGPDERARAWQRLGSEQFDVIVIGGGIVGAGA
Mvan_1620|M.vanbaalenii_PYR-1 MGHVIPAPGNGQTF----LGPDHRVRAWQRLGNEQFDVIVIGGGVVGAGA
MSMEG_1736|M.smegmatis_MC2_155 MTVPKPAPGDGQTF----LGPQQRAQAWEQLGNQQFDVVVIGGGVVGAGA
TH_1838|M.thermoresistible__bu VSHPILRPGHGPTL----LGPEQRQRAWDRLGSEQFDVVVIGGGVVGSGA
Rv3302c|M.tuberculosis_H37Rv MSNPIQAPDGGQGWPAAALGPAQRAVAWKRLGTEQFDVVVIGGGVVGSGC
Mb3330c|M.bovis_AF2122/97 MSNPIQAPDGGQGWPAAALGPAQRAVAWKRLGTEQFDVVVIGGGVVGSGC
MLBr_00713|M.leprae_Br4923 MTDLIQAPESEQTWPASALGPQQRAAAWERFGTEQFDVVVIGGGVVGSGC
MMAR_1225|M.marinum_M MSNPTDASQRDQTGPAGSLGPEQRALAWERLGAEQFDVVVIGGGVVGSGC
MUL_2667|M.ulcerans_Agy99 MSNPTDASQRDQTGPADSLGPEQRALAWERLGAEQFDVVVIGGGVVGSGC
MAV_4278|M.avium_104 MSDPIHALG-EPGSPASALGPRQRAAAWDRLGAEQFDVVVIGGGVVGSGC
MAB_3655c|M.abscessus_ATCC_199 MSDATSPSGSSGPT-YTFLGPEARSLNWERLGSEQFDVIVIGGGVVGAGS
: *** * *.::* :****:*****:**:*.
Mflv_4821|M.gilvum_PYR-GCK ALDAATRGLKVALVEARDFASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
Mvan_1620|M.vanbaalenii_PYR-1 ALDAATRGLKVALVEARDFASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
MSMEG_1736|M.smegmatis_MC2_155 ALDAATRGLKVALVEARDFASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
TH_1838|M.thermoresistible__bu ALDAATRGLKVALVEARDFASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
Rv3302c|M.tuberculosis_H37Rv ALDAATRGLKVALVEARDLASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
Mb3330c|M.bovis_AF2122/97 ALDAATRGLKVALVEARDLASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
MLBr_00713|M.leprae_Br4923 ALDAATRGLKVALVEARDLASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
MMAR_1225|M.marinum_M ALDAATRGLKVALIEARDLASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
MUL_2667|M.ulcerans_Agy99 ALDAATRGLKVALIEARDLASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
MAV_4278|M.avium_104 ALDAATRGLKVALVEARDFASGTSSRSSKMFHGGLRYLEQLEFGLVREAL
MAB_3655c|M.abscessus_ATCC_199 ALDAATRGLKVALVEARDFASGTSSRSSKMFHGGLRYLEQLEFGLVQEAL
*************:****:***************************:***
Mflv_4821|M.gilvum_PYR-GCK HERELSLTTLAPHLVKPLPFLFPLTNRVWERPYIAAGIFLYDQLGGAKSV
Mvan_1620|M.vanbaalenii_PYR-1 HERELSLTTLAPHLVKPLPFLFPLTNRGWERPYIAAGIFLYDQLGGAKSV
MSMEG_1736|M.smegmatis_MC2_155 HERELSLTTLAPHLVKPLPFLFPLTNRWWERPYVAAGIFLYDQLGGAKSV
TH_1838|M.thermoresistible__bu HERELSLTTLAPHLVKPLPFLYPLTRRWWERPYVALGIFLYDQLGGARSL
Rv3302c|M.tuberculosis_H37Rv YERELSLTTLAPHLVKPLPFLFPLTKRWWERPYIAAGIFLYDRLGGAKSV
Mb3330c|M.bovis_AF2122/97 YERELSLTTLAPHLVKPLPFLFPLTKRWWERPYIAAGIFLYDRLGGAKSV
MLBr_00713|M.leprae_Br4923 YERELSLTTLAPHLVKPLPFLFPLTKRWWERPYIAAGIFLYDRLGGAKSV
MMAR_1225|M.marinum_M FERELSLTTLAPHLVKPLPFLFPLTKRGWERPYIAAGIFLYDRLGGAKSV
MUL_2667|M.ulcerans_Agy99 FERELSLTTLAPHLVKPLPFLFPLTKRGWERPYIAAGIFLYDRLGGAKSV
MAV_4278|M.avium_104 YERELSLTTLAPHLVKPMPFLFPLTNRLWERPYIAAGIFLYDRLGGAKSV
MAB_3655c|M.abscessus_ATCC_199 RERELSLSTLAPHLVKPLPFLFPLTSRWWERPYVAAGIFLYDQMGGAKSV
******:*********:***:*** * *****:* ******::***:*:
Mflv_4821|M.gilvum_PYR-GCK PPQKHLTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTAAHY
Mvan_1620|M.vanbaalenii_PYR-1 PAQKHLTRSGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTAAHY
MSMEG_1736|M.smegmatis_MC2_155 PAQKHLTKAGALRLAPGLKRSSLIGGIRYYDTVVDDARHTMTVARTAAHY
TH_1838|M.thermoresistible__bu PAQKHLTKAGALRLAPGLRRDSLIGGIRYYDTVVDDARHTLTVARTAAHY
Rv3302c|M.tuberculosis_H37Rv PAQRHFTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTAAHY
Mb3330c|M.bovis_AF2122/97 PAQRHFTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTAAHY
MLBr_00713|M.leprae_Br4923 PAQKHLTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTLTVARTAAHY
MMAR_1225|M.marinum_M PAQKHLTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTASHY
MUL_2667|M.ulcerans_Agy99 PAQKHLTRAGALRLSPGLKRSSLIGGIRYYDTVVDDARHTMTVARTASHY
MAV_4278|M.avium_104 PAQKHLTRAGALRLCPGLKRSALIGGIRYYDTIVDDARHTLTVARTAAHY
MAB_3655c|M.abscessus_ATCC_199 PAQKHLTRAGALRLAPGLKRNSLIGGIRYYDTVVDDARHTMTVARTAAHY
*.*:*:*::*****.***:*.:**********:*******:******:**
Mflv_4821|M.gilvum_PYR-GCK GAVVRTSTQVVALLREGDRVTGVQVRDSEDGRVTDVRGHVVVNATGVWTD
Mvan_1620|M.vanbaalenii_PYR-1 SAVVRTSTQVVALLREGDRVTGVRVRDSENGEVTEVRGHVVVNATGVWTD
MSMEG_1736|M.smegmatis_MC2_155 GAVVRSSTQVVALLREGDRVTGVRVRDSEDGSVTEVNGHVVVNATGVWTD
TH_1838|M.thermoresistible__bu GAVIRSSTQVVALLREGDRVTGVTVRDSETGAVTDVHGHVVVNATGVWTD
Rv3302c|M.tuberculosis_H37Rv GAVVRCSTQVVALLREGDRVIGVGVRDSENGAVAEVRGHVVVNATGVWTD
Mb3330c|M.bovis_AF2122/97 GAVVRCSTQVVALLREGDRVIGVGVRDSENGAVAEVRGHVVVNATGVWTD
MLBr_00713|M.leprae_Br4923 GAVVRCSTQVVALLREGDRVIGVRVRDSEDGAVTEIRGHVVVNATGVWTD
MMAR_1225|M.marinum_M GAVVRCSTQVVALLREGDRVVGVRVRDSEDGSITEVRGHVVVNATGVWTD
MUL_2667|M.ulcerans_Agy99 GAVVRCSTQVVALLREGDRVVGVRVRDSEDGSITEVRGHVVVNATGVWTD
MAV_4278|M.avium_104 GVVVRNSTQVVALLREGDRVTGVRVRDSEDGSVTEVRGHVVVNATGVWTD
MAB_3655c|M.abscessus_ATCC_199 GAVVRASTQVVSLLREGDRVTGVLVRDTESGAETEVRGHVVINATGVWTD
..*:* *****:******** ** ***:* * :::.****:********
Mflv_4821|M.gilvum_PYR-GCK EIQALSKQRGRFRVRASKGVHIVVPRDRIVSEVAIILRTEKSVLFVIPWG
Mvan_1620|M.vanbaalenii_PYR-1 EIQALSKQRGRFRVRASKGVHIVVPRDRIVSEVAIILRTEKSVLFVIPWG
MSMEG_1736|M.smegmatis_MC2_155 EIQALSKQRGRFRVRASKGVHIVVPRDRIVSEVAIILRTEKSVLFVIPWG
TH_1838|M.thermoresistible__bu EIQALSKQRGRFRVRASKGVHIVVPRDRIVSEVAIILRTEKSVLFVIPWG
Rv3302c|M.tuberculosis_H37Rv EIQALSKQRGRFQVRASKGVHVVVPRDRIVSDVAMILRTEKSVMFVIPWG
Mb3330c|M.bovis_AF2122/97 EIQALSKQRGRFQVRASKGVHVVVPRDRIVSDVAMILRTEKSVMFVIPWG
MLBr_00713|M.leprae_Br4923 EIQALSKQRGRFQVRVSKGVHVVVPRDRIVSDVAMILRTKKSVMFIIPWG
MMAR_1225|M.marinum_M EIQALSKQRGRFQVRASKGVHVVVPRDRIVSDVAMILRTEKSVMFIIPWG
MUL_2667|M.ulcerans_Agy99 EIQALSKQRGRFQVRASKGVHVVVPRDRIVSDVAMILRTEKSVMFIIPWG
MAV_4278|M.avium_104 EIQALSKQRGRFQVRASKGVHVVVPRDRIVSDVAIILRTEKSVMFVIPWG
MAB_3655c|M.abscessus_ATCC_199 EIQALSKTRGRFRVTASKGVHIVVPRDRIVSEVAIILRTEKSVLFVIPWG
******* ****:* .*****:*********:**:****:***:*:****
Mflv_4821|M.gilvum_PYR-GCK THWIIGTTDTDWNLDLAHPAATKADIDYILGHVNKVLATPLTHDDIDGVY
Mvan_1620|M.vanbaalenii_PYR-1 THWIIGTTDTDWHLDLAHPAATKADIDYILGHVNKVLATPLTHDDIDGVY
MSMEG_1736|M.smegmatis_MC2_155 THWIIGTTDTDWNLDLAHPAATKADIDYILGHVNTVLATPLTHDDIDGVY
TH_1838|M.thermoresistible__bu THWIIGTTDTDWNLDLAHPAATKADIDYILGQVNRALATPLTHADIDGVY
Rv3302c|M.tuberculosis_H37Rv SHWIIGTTDTDWNLDLAHPAATKADIDYILGTVNAVLATPLTHADIDGVY
Mb3330c|M.bovis_AF2122/97 SHWIIGTTDTDWNLDLAHPAATKADIDYILGTVNAVLATPLTHADIDGVY
MLBr_00713|M.leprae_Br4923 NHWIIGTTDTDWNLDLAHPAATKADIDYILQTVNTVLATPLTHADIDGVY
MMAR_1225|M.marinum_M SHWIIGTTDTDWNLDLAHPAATKADIDYILGTVNAVLATPLTHADIDGVY
MUL_2667|M.ulcerans_Agy99 SHWIIGTTDTDWNLDLAHPAATKADIDYILGTVNAVLATPLTHADIDGVY
MAV_4278|M.avium_104 SHWIIGTTDTDWDLDLAHPAATKADIDYILSTVNTVLAIPLTHADIDGVY
MAB_3655c|M.abscessus_ATCC_199 NHWIIGTTDTEWNLDLAHPAATKVDIDYILDHVNTVLATPLTHNDIDGVY
.*********:*.**********.****** ** .** **** ******
Mflv_4821|M.gilvum_PYR-GCK AGLRPLLAGESEETSKLSREHAVAVPSPGLVAIAGGKYTTYRVMAEDAID
Mvan_1620|M.vanbaalenii_PYR-1 AGLRPLLAGESEETSKLSREHAVAVPAPGLVAIAGGKYTTYRVMAEDAID
MSMEG_1736|M.smegmatis_MC2_155 AGLRPLLAGESEETSKLSREHAVAVPAPGLVAIAGGKYTTYRVMAADAID
TH_1838|M.thermoresistible__bu AGLRPLLAGESEETSKLSREHAVAVPAPGLIAIAGGKYTTYRVMGEDAID
Rv3302c|M.tuberculosis_H37Rv AGLRPLLAGESDDTSKLSREHAVAVPAAGLVAIAGGKYTTYRVMAADAID
Mb3330c|M.bovis_AF2122/97 AGLRPLLAGESDDTSKLSREHAVAVPAAGLVAIAGGKYTTYRVMAADAID
MLBr_00713|M.leprae_Br4923 AGLRPLLAGESDDTSKLTREHAVAVPVAGLVAIAGGKYTTYRVMAADAID
MMAR_1225|M.marinum_M AGLRPLLAGESDDTSKLSREHAVAVPAAGLVAIAGGKYTTYRVMAADAID
MUL_2667|M.ulcerans_Agy99 AGLRPLLAGESDDTSKLSREHAVAVPAAGLVAIAGGKYTTYRVMAADAID
MAV_4278|M.avium_104 AGLRPLLAGESEETSKLSREHAVAVPAPGLVAIAGGKYTTYRVMAADAID
MAB_3655c|M.abscessus_ATCC_199 AGLRPLLAGENDDTSKLSREHAVAVAAPGLVAIAGGKYTTYRVMASDAVD
**********.::****:*******. .**:*************. **:*
Mflv_4821|M.gilvum_PYR-GCK AAAEFIPTRVAPSITEKVPLMGADGYFALINQTQSVGTYYDLHPYRVRHL
Mvan_1620|M.vanbaalenii_PYR-1 AAAEFIPTRVAPSITERVPLMGADGYFALINQTQSVGAHYGLHPYRVRHL
MSMEG_1736|M.smegmatis_MC2_155 AAAEFVPTRVAPSITEKVPLMGADGYFALINQTEHVGRQYGLHPYRVRHL
TH_1838|M.thermoresistible__bu AAAEFIPARVAPSITEKVPLLGADGYFALINQTETVGAHYGLHPYRVRHL
Rv3302c|M.tuberculosis_H37Rv AAVQFIPARVAPSITEKVSLLGADGYFALVNQAEHVGALQGLHPYRVRHL
Mb3330c|M.bovis_AF2122/97 AAVQFIPARVAPSITEKVSLLGADGYFALVNQAEHVGALQGLHPYRVRHL
MLBr_00713|M.leprae_Br4923 AAVAFVPARVAPSITEKVGLLGADGYFALINQVEHVAALQGLHPYRVRHL
MMAR_1225|M.marinum_M AAVQFVPARVAPSITEKVSLLGADGYFALVNQVEHVGEMVGLHPYRVRHL
MUL_2667|M.ulcerans_Agy99 AAVQFVPARVAPSITEKVSLLGADGYFALVNQVEHVGEMVGLHPYRVRHL
MAV_4278|M.avium_104 AAAQFIPARVAPSITEKVSLLGADGYFALINQAEHVAQLRGLHPYRVRHL
MAB_3655c|M.abscessus_ATCC_199 MASEFVPARVAPSITEKVPLLGADGYFALINQTEHVGETYGLHPYRVRHL
* *:*:********:* *:********:**.: *. .*********
Mflv_4821|M.gilvum_PYR-GCK LDRYGSLIGEVLKLAED----NPDLLNPITEAPVYLKVEAAYAAAAEGAL
Mvan_1620|M.vanbaalenii_PYR-1 LDRYGSLIGEVLKLASDLAAVRPDLLEPITDAPVYLKVEAAYAAAAEGAL
MSMEG_1736|M.smegmatis_MC2_155 LDRYGSLIGEVLALADG----KPELLEPITEAPVYLKVEAAYAAAAESAL
TH_1838|M.thermoresistible__bu LDRYGSLIGEVLQMAGRDGAGRPDLLEPITDAPVYLKVEAWYAAAAEGAL
Rv3302c|M.tuberculosis_H37Rv LDRYGSLISDVLAMAAS----DPSLLSPITEAPGYLKVEAAYAAAAEGAL
Mb3330c|M.bovis_AF2122/97 LDRYGSLISDVLAMAAS----DPSLLSPITEAPGYLKVEAAYAAAAEGAL
MLBr_00713|M.leprae_Br4923 LDRYGALIGDVLALAAE----APDLLSPIQEAPGYLKVEARYAVTAEGAL
MMAR_1225|M.marinum_M LDRYGSLINEVLELAAD----DPDLLTPIKEAPGYLKVEALYAVTAEGAL
MUL_2667|M.ulcerans_Agy99 LDRYGSLINEVLELAAD----DPDLLTPIKEAPGYLKVEALYAVTAEGAL
MAV_4278|M.avium_104 LDRYGSLIGDVLALAAD----RPDLLNPITEAPGYLRVEALYAVTAEGAL
MAB_3655c|M.abscessus_ATCC_199 LDRYGSLIGEVLACAGT----DHSLLQPITKAPMYLKVEARYAVAAEGAL
*****:**.:** * .** ** .** **:*** **.:**.**
Mflv_4821|M.gilvum_PYR-GCK HLEDILARRMRISIEYPHRGVDCAREVAEVVAPILGWSPEDVDREVSTYL
Mvan_1620|M.vanbaalenii_PYR-1 HLEDILARRMRISIEYPHRGVDCAREVAEVVAPLLGWSAEDVDREVDTYL
MSMEG_1736|M.smegmatis_MC2_155 HLEDILARRMRISIEYPHRGVECAREVAEVVAPILGWSVEDVDREVATYA
TH_1838|M.thermoresistible__bu HLEDILARRMRISIEYPHRGVDCAREVAEVVAPVLGWSDADIEREVDTYC
Rv3302c|M.tuberculosis_H37Rv HLEDILARRMRISIEYPHRGVDCAREVAEVVAPVLGWTAADIDREVANYM
Mb3330c|M.bovis_AF2122/97 HLEDILARRMRISIEYPHRGVDCAREVAEVVAPVLGWTAADIDREVANYM
MLBr_00713|M.leprae_Br4923 HLEDILARRMRVSIEYPHRGVACAREVADVVAPVLGWTAEDIDREVATYN
MMAR_1225|M.marinum_M HLEDILARRMRVSIEYPHRGVDCAREVADVVAPVLGWTVEDIEREVANYK
MUL_2667|M.ulcerans_Agy99 HLEDILARRMRVSIEYPHRGVDCAREVADVVAPVLGWTVEDIEREVANYK
MAV_4278|M.avium_104 HLEDILARRMRISIEYSHRGVDCAREVADVVAPVLGWTAGDVEREVANYT
MAB_3655c|M.abscessus_ATCC_199 HLEDILARRMRIAIEYPHRGVDCAQEVAELVAPVLGWSAEDIVREVDTYR
***********::***.**** **:***::***:***: *: *** .*
Mflv_4821|M.gilvum_PYR-GCK ARVEAEVQSQTQPDDASADALRAAAPEARAEILEPVPLN---------
Mvan_1620|M.vanbaalenii_PYR-1 ARVEAEVLSQTQPDDESADALRAAAPEARAEILEPVPLK---------
MSMEG_1736|M.smegmatis_MC2_155 ARVDAEVRSQQQPDDESADALRQAAPEARAEILEPVPLN---------
TH_1838|M.thermoresistible__bu ARVEAEVRSQQQPDDESADALRQAAPEARAMILEPVPLD---------
Rv3302c|M.tuberculosis_H37Rv ARVEAEVLSQAQPDDVSADMLRASAPEARAEILEPVPLD---------
Mb3330c|M.bovis_AF2122/97 ARVEAEVLSQAQPDDVSADMLRASAPEARAEILEPVPLD---------
MLBr_00713|M.leprae_Br4923 ARVEAEVLSQAQPDDVSADMLRASAPEARTKIIEPVSLT---------
MMAR_1225|M.marinum_M ARVEAEVLSQAQPDDVSADMLRASAPEARAEILEPVPLN---------
MUL_2667|M.ulcerans_Agy99 ARVEAEVLSQAQPDDVSADMLRASAPEARAEILEPVPLN---------
MAV_4278|M.avium_104 ARVQAEILSQAQPDDVSADELRASAPEARAEILEPVPLN---------
MAB_3655c|M.abscessus_ATCC_199 ARVDAEVRSQRQPDDASADALRVAAPEARSFIVEPVTETGTVLSPATK
***:**: ** **** *** ** :*****: *:***.