For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRRSASTCGWKTPTRRGTSRPSDSKTLILELPDERAVAIVPVPSKLSLKAAGGPRGAQSGHG
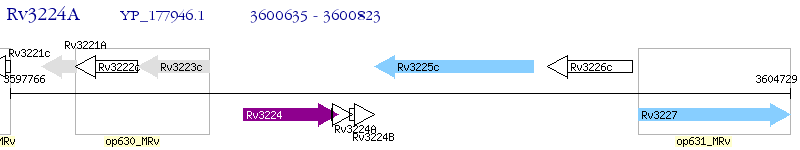
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3224A | - | - | 100% (62) | hypothetical protein Rv3224A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3252 | - | 1e-31 | 100.00% (62) | hypothetical protein Mb3252 |
| M. gilvum PYR-GCK | Mflv_4690 | - | 2e-05 | 60.61% (33) | ybaK/ebsC protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1332 | - | 8e-07 | 69.70% (33) | hypothetical protein MMAR_1332 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1913 | ybaK | 2e-05 | 62.50% (32) | YbaK/ebsC protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2547 | - | 9e-07 | 66.67% (33) | hypothetical protein MUL_2547 |
| M. vanbaalenii PYR-1 | Mvan_1777 | - | 3e-05 | 60.61% (33) | ybaK/ebsC protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4690|M.gilvum_PYR-GCK MTGSDSRSAGQNNSGRGRACSRAAAAYAVPVARAATPAIAALVAAGVAHD
Mvan_1777|M.vanbaalenii_PYR-1 ------MAAG---------VVVPCPAYAVPVARAATPAVAALVAAGVAHE
MSMEG_1913|M.smegmatis_MC2_155 ------------------------------MARAATPAIAALVAAGIQHD
MMAR_1332|M.marinum_M ------------------------------MARAATPGVAALIKAGVTHE
MUL_2547|M.ulcerans_Agy99 --------------------------------------------------
Rv3224A|M.tuberculosis_H37Rv ------------------------------MRRSAS-------TCGWKTP
Mb3252|M.bovis_AF2122/97 ------------------------------MRRSAS-------TCGWKTP
Mflv_4690|M.gilvum_PYR-GCK VVQYHHDPRTDSFGAEAVSELAGAGGLAPEQIFKTLVVSLPRGLGVAVLP
Mvan_1777|M.vanbaalenii_PYR-1 VLQYRHDPRSESFGEEAVKELAQAGGLAPAQILKTLVVALPKGLGVAVLP
MSMEG_1913|M.smegmatis_MC2_155 VVRYHHDPRNDSFGEEAAAQLAADG-YVAEQVFKTLVIALPKGLAVAVVP
MMAR_1332|M.marinum_M LLHF-----------EAVTDLSDRADIAPEQVFKTLIIALPGELAVAVVP
MUL_2547|M.ulcerans_Agy99 -------------------------------MFKTLVIALPGEFAVAVVP
Rv3224A|M.tuberculosis_H37Rv ---------------------TRRGTSRPSDS-KTLILELPDERAVAIVP
Mb3252|M.bovis_AF2122/97 ---------------------TRRGTSRPSDS-KTLILELPDERAVAIVP
***:: ** .**::*
Mflv_4690|M.gilvum_PYR-GCK VPSKLSLKAAAAALGAAKATMADPAAAQRSSGYVVGGISPLGQRKALPTV
Mvan_1777|M.vanbaalenii_PYR-1 VPSKLSLKAAAAALGAARATMADPADAQRSSGYVVGGISPLGQRKALPTV
MSMEG_1913|M.smegmatis_MC2_155 VPTKLSLKAAAAALGVARAEMADPAAAQRSTGYVVGGISPLGQRKPLPTV
MMAR_1332|M.marinum_M VPSRLSLKAAASALGAAKASMAQPADAERTTGYVVGGISPFGQRRKLRTV
MUL_2547|M.ulcerans_Agy99 VPSRLSLKAAASALGAAKASMAEPADAERTTGYVVGGISPFGQRRKLRTV
Rv3224A|M.tuberculosis_H37Rv VPSKLSLKAAG-----------GPRGAQSGHG------------------
Mb3252|M.bovis_AF2122/97 VPSKLSLKAAG-----------GPRGAQSGHG------------------
**::******. * *: *
Mflv_4690|M.gilvum_PYR-GCK VDASALNWDRILCSAGKRGWDVSLDPRDLVRLTNAVTADICAL-
Mvan_1777|M.vanbaalenii_PYR-1 VDSSAMQWDRVLCSAGKRGWDVALDPRDLVRLTNAVTADICAP-
MSMEG_1913|M.smegmatis_MC2_155 IDASALRFDKVLCSAGKRGLDIVLAPADLIRATDAVTAEIAVGA
MMAR_1332|M.marinum_M VDSSALTWDRVLCSAGKRHWDVAVPPADLVRLTDALTADIRA--
MUL_2547|M.ulcerans_Agy99 VDSSALTWDRVLCSAGKRHWDVAVPPADLVRLTDALTADIRA--
Rv3224A|M.tuberculosis_H37Rv --------------------------------------------
Mb3252|M.bovis_AF2122/97 --------------------------------------------