For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARAATPAIAALVAAGIQHDVVRYHHDPRNDSFGEEAAAQLAADGYVAEQVFKTLVIALPKGLAVAVVPVP TKLSLKAAAAALGVARAEMADPAAAQRSTGYVVGGISPLGQRKPLPTVIDASALRFDKVLCSAGKRGLDI VLAPADLIRATDAVTAEIAVGA*
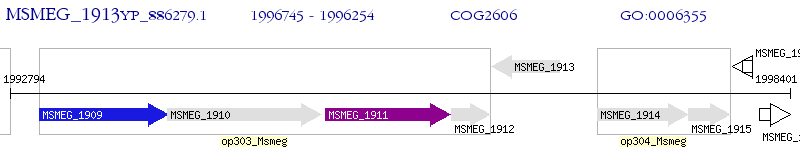
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1913 | ybaK | - | 100% (163) | YbaK/ebsC protein |
| M. smegmatis MC2 155 | MSMEG_5671 | - | 5e-05 | 23.78% (143) | YbaK/prolyl-tRNA synthetase associated region |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3253 | - | 1e-12 | 68.09% (47) | hypothetical protein Mb3253 |
| M. gilvum PYR-GCK | Mflv_4690 | - | 8e-65 | 75.62% (160) | ybaK/ebsC protein |
| M. tuberculosis H37Rv | Rv3224B | - | 1e-12 | 68.09% (47) | hypothetical protein Rv3224B |
| M. leprae Br4923 | MLBr_00799 | - | 2e-25 | 62.35% (85) | hypothetical protein MLBr_00799 |
| M. abscessus ATCC 19977 | MAB_3544 | - | 5e-54 | 64.81% (162) | hypothetical protein MAB_3544 |
| M. marinum M | MMAR_1332 | - | 1e-46 | 60.00% (160) | hypothetical protein MMAR_1332 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2547 | - | 6e-39 | 70.64% (109) | hypothetical protein MUL_2547 |
| M. vanbaalenii PYR-1 | Mvan_1777 | - | 8e-62 | 72.50% (160) | ybaK/ebsC protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4690|M.gilvum_PYR-GCK MTGSDSRSAGQNNSGRGRACSRAAAAYAVPVARAATPAIAALVAAGVAHD
Mvan_1777|M.vanbaalenii_PYR-1 ------MAAG---------VVVPCPAYAVPVARAATPAVAALVAAGVAHE
MSMEG_1913|M.smegmatis_MC2_155 ------------------------------MARAATPAIAALVAAGIQHD
MAB_3544|M.abscessus_ATCC_1997 --------------------------MGSKAVKAATPAVAALIAGGAEYE
MMAR_1332|M.marinum_M ------------------------------MARAATPGVAALIKAGVTHE
MUL_2547|M.ulcerans_Agy99 --------------------------------------------------
Mb3253|M.bovis_AF2122/97 --------------------------------------------------
Rv3224B|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00799|M.leprae_Br4923 ---------------------------------MQFDTVNNLSRNTLTPR
Mflv_4690|M.gilvum_PYR-GCK VVQYHHDPRTDSFGAEAVSELAGAGGLAPEQIFKTLVVSLPRG--LGVAV
Mvan_1777|M.vanbaalenii_PYR-1 VLQYRHDPRSESFGEEAVKELAQAGGLAPAQILKTLVVALPKG--LGVAV
MSMEG_1913|M.smegmatis_MC2_155 VVRYHHDPRNDSFGEEAAAQLAADG-YVAEQVFKTLVIALPKG--LAVAV
MAB_3544|M.abscessus_ATCC_1997 LLPYEHRAGEHAFGEEAVRALAGSG-LVPEQIFKTLLISLDRGSDLAVAV
MMAR_1332|M.marinum_M LLHF-----------EAVTDLSDRADIAPEQVFKTLIIALPGE--LAVAV
MUL_2547|M.ulcerans_Agy99 -------------------------------MFKTLVIALPGE--FAVAV
Mb3253|M.bovis_AF2122/97 --------------------------------------------------
Rv3224B|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00799|M.leprae_Br4923 LSRYS----------------------------KTLIIALPRE--LAIAI
Mflv_4690|M.gilvum_PYR-GCK LPVPSKLSLKAAAAALGAAKATMADPAAAQRSSGYVVGGISPLGQRKALP
Mvan_1777|M.vanbaalenii_PYR-1 LPVPSKLSLKAAAAALGAARATMADPADAQRSSGYVVGGISPLGQRKALP
MSMEG_1913|M.smegmatis_MC2_155 VPVPTKLSLKAAAAALGVARAEMADPAAAQRSTGYVVGGISPLGQRKPLP
MAB_3544|M.abscessus_ATCC_1997 IPVPATLSLKAAAAALGRAKATMADPAAAERSSGYVVGGISPLGQRKQLP
MMAR_1332|M.marinum_M VPVPSRLSLKAAASALGAAKASMAQPADAERTTGYVVGGISPFGQRRKLR
MUL_2547|M.ulcerans_Agy99 VPVPSRLSLKAAASALGAAKASMAEPADAERTTGYVVGGISPFGQRRKLR
Mb3253|M.bovis_AF2122/97 -----------------MPKAAMAKPAAAEQATGYVVGGISPFGQRKRLR
Rv3224B|M.tuberculosis_H37Rv -----------------MPKAAMAKPAAAEQATGYVVGGISPFGQRKRLR
MLBr_00799|M.leprae_Br4923 LSAPSRLSLKETAAAPGVTKANTAEPAAAQRATSYVVGGVFPSGQWKRLR
.:* *.** *::::.*****: * ** : *
Mflv_4690|M.gilvum_PYR-GCK TVVDASALNWDRILCSAGKRGWDVSLDPRDLVRLTNAVTADICAL---
Mvan_1777|M.vanbaalenii_PYR-1 TVVDSSAMQWDRVLCSAGKRGWDVALDPRDLVRLTNAVTADICAP---
MSMEG_1913|M.smegmatis_MC2_155 TVIDASALRFDKVLCSAGKRGLDIVLAPADLIRATDAVTAEIAVGA--
MAB_3544|M.abscessus_ATCC_1997 TVIDASALTWERILCSAGKRGLQMALAPEDLVRLTGAVTAPVIAG---
MMAR_1332|M.marinum_M TVVDSSALTWDRVLCSAGKRHWDVAVPPADLVRLTDALTADIRA----
MUL_2547|M.ulcerans_Agy99 TVVDSSALTWDRVLCSAGKRHWDVAVPPADLVRLTDALTADIRA----
Mb3253|M.bovis_AF2122/97 TVVDVSALSWDRVL-----RCRQTALGRHGGPAGPDHLD--QRDHR--
Rv3224B|M.tuberculosis_H37Rv TVVDVSALSWDRVL-----RCRQTALGRHGGPAGPDHLD--QRDHR--
MLBr_00799|M.leprae_Br4923 TVVDTSALSWDRVLCNAGKRHRLLAPGCRGEPGYVRHPSGWRSAERCW
**:* **: ::::* * .