For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STLGDLLAEHTVLPGSAVDHLHAVVGEWQLLADLSFADYLMWVRRDDGVLVCVAQCRPNTGPTVVHTDAV GTVVAANSMPLVAATFSGGVPGREGAVGQQNSCQHDGHSVEVSPVRFGDQVVAVLTRHQPELAARRRSGH LETAYRLCATDLLRMLAEGTFPDAGDVAMSRSSPRAGDGFIRLDVDGVVSYASPNALSAYHRMGLTTELE GVNLIDATRPLISDPFEAHEVDEHVQDLLAGDGKGMRMEVDAGGATVLLRTLPLVVAGRNVGAAILIRDV TEVKRRDRALISKDATIREIHHRVKNNLQTVAALLRLQARRTSNAEGREALIESVRRVSSIALVHDALSM SVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGDLGVLDSDRATALIMVITELVQNAIEHAFDPAAAE GSVTIRAERSARWLDVVVHDDGLGLPQGFSLEKSDSLGLQIVRTLVSAELDGSLGMRDARERGTDVVLRV PVGRRGRLML*
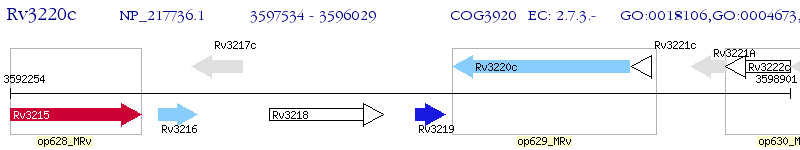
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3220c | - | - | 100% (501) | PROBABLE TWO COMPONENT SENSOR KINASE |
| M. tuberculosis H37Rv | Rv2027c | - | 2e-05 | 26.47% (170) | histidine kinase response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3246c | - | 0.0 | 99.80% (501) | two component sensor kinase |
| M. gilvum PYR-GCK | Mflv_4686 | - | 0.0 | 77.56% (499) | signal transduction histidine kinase |
| M. leprae Br4923 | MLBr_00803 | - | 0.0 | 81.84% (501) | putative two-component system sensor kinase |
| M. abscessus ATCC 19977 | MAB_3540c | - | 0.0 | 72.85% (501) | two component sensor kinase |
| M. marinum M | MMAR_1337 | - | 0.0 | 85.40% (500) | sensor kinase from two-component regulatory system |
| M. avium 104 | MAV_4168 | - | 0.0 | 84.43% (501) | sensor histidine kinase |
| M. smegmatis MC2 155 | MSMEG_1918 | - | 0.0 | 75.70% (498) | sensor histidine kinase |
| M. thermoresistible (build 8) | TH_0668 | - | 0.0 | 76.51% (498) | PROBABLE TWO COMPONENT SENSOR KINASE |
| M. ulcerans Agy99 | MUL_2542 | - | 0.0 | 85.20% (500) | sensor kinase from two component regulatory system |
| M. vanbaalenii PYR-1 | Mvan_1781 | - | 0.0 | 77.51% (498) | signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4686|M.gilvum_PYR-GCK MSTLGDLLAEHTILPGNAVDHLHAVVGEWQLLADLSFADYLMWVRRDDGH
Mvan_1781|M.vanbaalenii_PYR-1 MSTLGDLLAEHTVLPGNAVDHLHAVVGEWQLLADLSFADYLMWVRRDDGM
MSMEG_1918|M.smegmatis_MC2_155 MSTLGDLLAEHTMLPGSAVDHLHAVVGEWQMLSDLSFADYLMWVRRDDGV
TH_0668|M.thermoresistible__bu MSTLGDLLAEHTVLPGAAVDHLHAVVGEWQLLADMSFADYLMWVRRDDGR
Rv3220c|M.tuberculosis_H37Rv MSTLGDLLAEHTVLPGSAVDHLHAVVGEWQLLADLSFADYLMWVRRDDGV
Mb3246c|M.bovis_AF2122/97 MSTLGDLLAEHTVLPGSAVDHLHAVVGEWQLLADLSFADYLMWVRRNDGV
MMAR_1337|M.marinum_M MSTLGNLLAENTMLPGNAVDHLHAVVGEWQLLADLSFADYLMWVARDDGA
MUL_2542|M.ulcerans_Agy99 MSTLGNLLAENTMLPGNAVDHLHAVVGEWQLLADLSFADYLMWVARDDGA
MAV_4168|M.avium_104 MSTLGDLLAEHTMLPGNAVDHLHAVVGEWQLLADLSFADYLMWVCRDDGM
MLBr_00803|M.leprae_Br4923 MSTLGDLLAEHTVLPGNAVGHLHAVVGEWQLLADLSFADYLMWVSRDDGV
MAB_3540c|M.abscessus_ATCC_199 MSTLGDLLAEHTMLPGDAVDHLHAVVAEWQLLADLSFADYLMWVHRDDDA
*****:****:*:*** **.******.***:*:*:********* *:*.
Mflv_4686|M.gilvum_PYR-GCK LVCVAQVRPNTAPTVLLADAVGTLAEADDIPIVAAAFRSGDIGRATVDGE
Mvan_1781|M.vanbaalenii_PYR-1 LVCVAQVRPNTAATVLLADAVGTLAEAEDLPIVAEAFQSGTIGRATVEGE
MSMEG_1918|M.smegmatis_MC2_155 LVCVAQIRPNTAPTVLLADSVGKLAGPDDLPVVTMAFESGAIGRGSDKAG
TH_0668|M.thermoresistible__bu LVCVAQVRPNTAHTVLLTDAVGTIAEPDEMPLVSAAFETGAIGRESGAEQ
Rv3220c|M.tuberculosis_H37Rv LVCVAQCRPNTGPTVVHTDAVGTVVAANSMPLVAATFSGGVPGREGAVGQ
Mb3246c|M.bovis_AF2122/97 LVCVAQCRPNTGPTVVHTDAVGTVVAANSMPLVAATFSGGVPGREGAVGQ
MMAR_1337|M.marinum_M LVCVAQCRPNTAPTVLQTDAVGSVVEADRLPLVAETFASGSARRDSDADQ
MUL_2542|M.ulcerans_Agy99 LVCVAQCRPNTAPTVLQTDAVGSVVEADRLPLVAETFASGSARRDSDADQ
MAV_4168|M.avium_104 LVCVAQCRPNTAPTVLQTDAVGTVVASERLTLVAETFASGAAKADDVAAQ
MLBr_00803|M.leprae_Br4923 LVCVAQCRPHTAPTVLQTDAVGSVASADMLPLVTEMFTSGAVQHEDAAGQ
MAB_3540c|M.abscessus_ATCC_199 FVCVAQCRPNTAPTIIDNDAVGTIAKIADVPLVEAAFASGAIERESDSDL
:***** **:*. *:: *:**.:. :.:* * *
Mflv_4686|M.gilvum_PYR-GCK FGA---PGLNVEAVPVRHGSDVVAVLT-HQTSLAPR-QASPLEAAYVDCA
Mvan_1781|M.vanbaalenii_PYR-1 FGS---PGLNVEAVPVRYRNDVVAVLT-HQTSLAPR-QASPLEAAYVDCA
MSMEG_1918|M.smegmatis_MC2_155 QGSRTRPGLDVEAVPVRHQGQVVAVLT-HQTALAERRLTSPLEGAYLDCA
TH_0668|M.thermoresistible__bu RLL---PGLNVEAVPVRYGDSVVAVLT-HQTALEARRTASPLEGAYLDCA
Rv3220c|M.tuberculosis_H37Rv QNSCQHDGHSVEVSPVRFGDQVVAVLTRHQPELAARRRSGHLETAYRLCA
Mb3246c|M.bovis_AF2122/97 QNSCQHDGHSVEVSPVRFGDQVVAVLTRHQPELAARRRSGHLETAYRLCA
MMAR_1337|M.marinum_M QHLGQLPGPSIEASPVRYGDQVVAVLTQHQTAVAAERASGHLESAYRDCA
MUL_2542|M.ulcerans_Agy99 QHLGQLPGPSIEASPVRYGDQVVAVLTQHQTAVAAERASGHLESAYRDCA
MAV_4168|M.avium_104 QDS-WLPGTHVEASPVRYGGHVVAVLTRHQTAVAADRASGQLEIAYRECA
MLBr_00803|M.leprae_Br4923 QYSWLPFGRNVAASPVRYGDQVVAVLTQHQVELAARRRPGNLEIAYLACA
MAB_3540c|M.abscessus_ATCC_199 TEH--ASELNVEAVPVRCGDRTVAVLT-HQTALASRLKKGTLERAYLECA
: . *** . .***** ** : . ** ** **
Mflv_4686|M.gilvum_PYR-GCK GDLLHMLSEGTFPNVGDLAMSRSSPRVGDGFIRLDEAGVVTFASPNAISA
Mvan_1781|M.vanbaalenii_PYR-1 GDLLHMLSEGTFPNVGDLAMSRSSPRAGDGFIRLDEAGVVTFASPNAISA
MSMEG_1918|M.smegmatis_MC2_155 NDLLHMLAEGTFPNIGDLAMSRSSPRVGDGFIRLNEGGEVVFASPNAISA
TH_0668|M.thermoresistible__bu EDLLLMLSEGTFPNVGDLPVSRSSPRVGDGFIRLDRTGGVVYASPNALSA
Rv3220c|M.tuberculosis_H37Rv TDLLRMLAEGTFPDAGDVAMSRSSPRAGDGFIRLDVDGVVSYASPNALSA
Mb3246c|M.bovis_AF2122/97 TDLLRMLAEGTFPDAGDVAMSRSSPRAGDGFIRLDVDGVVSYASPNALSA
MMAR_1337|M.marinum_M TDLLQMLAEGTFPDVGDVAMSRSTPRAGDGFIRLDVDGVAIYASPNALSA
MUL_2542|M.ulcerans_Agy99 TDLLQMLAEGTFPDVGDVAMSRSTPRAGDGFIRLDVDGVAIYASPNALSA
MAV_4168|M.avium_104 ADLVHMLAEGTFPDVGDVAMSRSTPRAGDGFIRLDVNGVVAYASPNALSA
MLBr_00803|M.leprae_Br4923 TDLLHMLAEGTFPDVGDVAVSHSTPRAGDGFIRLDRNGVVAFASPNALSA
MAB_3540c|M.abscessus_ATCC_199 GDLLLMLTEGTFPNVADQAMSRSSPRVGDGFIRLDVSGGVKYASPNALSA
**: **:*****: .* .:*:*:**.*******: * . :*****:**
Mflv_4686|M.gilvum_PYR-GCK YHRMGLSAELEGHNLVTVTRPLIADTFEAQELANHVRDSLAG-GSSMRME
Mvan_1781|M.vanbaalenii_PYR-1 YHRMGLTADLEGHNLVSITRPLISDPFEAQELANHVRDSLTG-GSSMRME
MSMEG_1918|M.smegmatis_MC2_155 YHRMGLNSELDGHNLVAITRPLISDPFEAQELANHIRDSLAG-GSSMRME
TH_0668|M.thermoresistible__bu YHRMGLTTELEGRNLVNVTRPLISDPFEAQELANHVTDSLAG-GSSMRME
Rv3220c|M.tuberculosis_H37Rv YHRMGLTTELEGVNLIDATRPLISDPFEAHEVDEHVQDLLAGDGKGMRME
Mb3246c|M.bovis_AF2122/97 YHRMGLTTELEGVNLIDATRPLISDPFEAHEVDEHVQDLLAGDGKGMRME
MMAR_1337|M.marinum_M YHRMGLTTELEGHELIEVTRPLISDPFEAEEVAEHVRDLLVG-GRSMRME
MUL_2542|M.ulcerans_Agy99 YHRMGLTTELEGHELIEVTRPLISDPFEAEEVAEHVRDLLVG-GRSMRME
MAV_4168|M.avium_104 YHRMGLTTELEGHNLIEITRPLISDSFEAQEMAEHVKDLLAG-GKSMRME
MLBr_00803|M.leprae_Br4923 FHRMGLTAELQGRNLVKVTRPLLSDPFEAQEVAKHVQDLLDG-GPSMRVE
MAB_3540c|M.abscessus_ATCC_199 YHRMGLASDLQGHNLMDVTKPLISDPFEGQELAAHIRDVLAG-GPSMRME
:***** ::*:* :*: *:**::*.**..*: *: * * * * .**:*
Mflv_4686|M.gilvum_PYR-GCK VDAGGATVLLRTLPLGVHGVGVGAAVLIRDVTEVKRRDRALLSKDATIRE
Mvan_1781|M.vanbaalenii_PYR-1 VDAGGATVLLRTLPLGVHGVAMGAAVLIRDVTEVKRRDRALLSKDATIRE
MSMEG_1918|M.smegmatis_MC2_155 VDANGAAILLRTLPLVAGGKSLGAAVLIRDVTEVKRRDRALLSKDATIRE
TH_0668|M.thermoresistible__bu VDAGSAAVLLRTLPLVVHGEPVGAVVLIRDVTEVKRRDRALLSKDATIRE
Rv3220c|M.tuberculosis_H37Rv VDAGGATVLLRTLPLVVAGRNVGAAILIRDVTEVKRRDRALISKDATIRE
Mb3246c|M.bovis_AF2122/97 VDAGGATVLLRTLPLVVAGRNVGAAILIRDVTEVKRRDRALISKDATIRE
MMAR_1337|M.marinum_M VDAGGATVLLRTLPLVVGGVSAGAAILIRDVTEVKRRDRALISKDATIRE
MUL_2542|M.ulcerans_Agy99 VDAGGATVLLRTLPLVVGGVSAGAAILIRDVTEVKRRDRALISKDATIRE
MAV_4168|M.avium_104 VDAGGATVLLRTLPLVVNGASAGAAVLIRDVTEVKRRDRALISKDATIRE
MLBr_00803|M.leprae_Br4923 VNAGGATVLLRTLPLVVHGSSVGAAVLVRDVTEVKRRDRALISKDATIRE
MAB_3540c|M.abscessus_ATCC_199 VDAGGAAVLIRTLPLIARGASEGALLLTRDVTEVKRRDRALLSKDATIRE
*:*..*::*:***** . * ** :* *************:********
Mflv_4686|M.gilvum_PYR-GCK IHHRVKNNLQTVAALLRLQARRTNNPEGREALMESVRRVSSIALVHDALS
Mvan_1781|M.vanbaalenii_PYR-1 IHHRVKNNLQTVAALLRLQARRTNNPEGREALMESVRRVSSIALVHDALS
MSMEG_1918|M.smegmatis_MC2_155 IHHRVKNNLQTVAALLRLQARRTSNEEAREALIESVRRVTSIALVHDALS
TH_0668|M.thermoresistible__bu IHHRVKNNLQTVASLLRLQARRTNNAEAREALLESVRRVSSIALVHDALS
Rv3220c|M.tuberculosis_H37Rv IHHRVKNNLQTVAALLRLQARRTSNAEGREALIESVRRVSSIALVHDALS
Mb3246c|M.bovis_AF2122/97 IHHRVKNNLQTVAALLRLQARRTSNAEGREALIESVRRVSSIALVHDALS
MMAR_1337|M.marinum_M IHHRVKNNLQTVAALLRLQARRTTNAEGREALIESVRRVSSIALVHDALS
MUL_2542|M.ulcerans_Agy99 IHHRVKNNLQTVAALLRLQARRTTNAEGREALTESVRRVSSIALVHDALS
MAV_4168|M.avium_104 IHHRVKNNLQTVAALLRLQARRTTNPEGREALIESVRRVSSIALVHDALS
MLBr_00803|M.leprae_Br4923 IHHRVKNNLQTVAALLRLQARRMANAEGREALIESVRRVSSIALVHDALS
MAB_3540c|M.abscessus_ATCC_199 IHHRVKNNLQTVAALLRLQARRTPNDEAREALFESVRRVSSIALVHDALS
*************:******** * *.**** ******:**********
Mflv_4686|M.gilvum_PYR-GCK MSVDEEVNLDEVIDRILPIMNDVAAVDSPIRINRVGDLGVLDADRATALT
Mvan_1781|M.vanbaalenii_PYR-1 MSVDEEVNLDQVIDRILPIMNDVAAVDSPIRINRVGDLGVLDADRATALI
MSMEG_1918|M.smegmatis_MC2_155 MSVDEEVNLDQVVDRILPIMNDVASVDTPIRINRVGNLGVLDADRATALI
TH_0668|M.thermoresistible__bu MSVDEEVNLDAVVDRILPIMNDVATVDTPIRIRRVGELGVLDADRATALI
Rv3220c|M.tuberculosis_H37Rv MSVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGDLGVLDSDRATALI
Mb3246c|M.bovis_AF2122/97 MSVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGDLGVLDSDRATALI
MMAR_1337|M.marinum_M MSVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGDLGVLDSDRATALI
MUL_2542|M.ulcerans_Agy99 MSVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGDLGVLDSDRATALI
MAV_4168|M.avium_104 MSVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGDLGVLDSDRATALI
MLBr_00803|M.leprae_Br4923 MSVDEQVNLDEVIDRILPIMNDVASVDRPIRINRVGNLGMLDSDRATALI
MAB_3540c|M.abscessus_ATCC_199 MSVDEEVNLDEVIDRIMPIMTDVATVDTKVRVSREGALGVLDSDRATPLI
*****:**** *:***:***.***:** :*: * * **:**:****.*
Mflv_4686|M.gilvum_PYR-GCK MVITELVQNAMEHAFDAETPQGCVTIRAQRSARWLDVIVHDDGRGLPEGF
Mvan_1781|M.vanbaalenii_PYR-1 MVITELVQNAIEHAFDATTPQGCVTIRAQRSARWLDVVVHDDGRGLPDGF
MSMEG_1918|M.smegmatis_MC2_155 MVITELVQNAIEHAFDAKTEQGCVTIKAERSARWLDVVVHDDGRGLPVGF
TH_0668|M.thermoresistible__bu MVITELAQNAIEHAFDRT--QGCVTIRAERSARFLDVVVHDDGRGLPAGF
Rv3220c|M.tuberculosis_H37Rv MVITELVQNAIEHAFDPAAAEGSVTIRAERSARWLDVVVHDDGLGLPQGF
Mb3246c|M.bovis_AF2122/97 MVITELVQNAIEHAFDPAAAEGSVTIRAERSARWLDVVVHDDGLGLPQGF
MMAR_1337|M.marinum_M MVITELVQNAIEHAFDP-AAQGAVTIRAERSARWLDVVVHDDGRGLPSGF
MUL_2542|M.ulcerans_Agy99 MVITELVQNAIEHAFDP-AAQGAVTIRAERSARWLDVVVHDDGRGLPSGF
MAV_4168|M.avium_104 MVITELVQNAIEHAFDPAAAEGSVTIRAERSARWLDVVVHDDGRGLPEGF
MLBr_00803|M.leprae_Br4923 MVITELVQNAIEHAFDPASQEGFVAISAERSARWLDVVVHDDGRGLPQGF
MAB_3540c|M.abscessus_ATCC_199 MVITELVQNAIEHAFDEG-AQGQVVIRAERSARWLDVVVHDDGRGLPEGF
******.***:***** :* *.* *:****:***:***** *** **
Mflv_4686|M.gilvum_PYR-GCK SLEKSDRLGLQIVRTLVSAELDGSLGMHEAAEGGTDVVLRVPIGRRVRVS
Mvan_1781|M.vanbaalenii_PYR-1 SLEKSDRLGLQIVRTLVSAELDGSLGMHAGAAGGTDVILRVPIGRRARPS
MSMEG_1918|M.smegmatis_MC2_155 SLEKSDRLGLQIVRTLVTAELDGSLGMHDVPSGGTDVVLRVPIGRRSRGA
TH_0668|M.thermoresistible__bu SLEKSDRLGLQIVRTLVTAELDGSISMNESPSGGTEVALRVPLGRRGRAA
Rv3220c|M.tuberculosis_H37Rv SLEKSDSLGLQIVRTLVSAELDGSLGMRDARERGTDVVLRVPVGRRGRLM
Mb3246c|M.bovis_AF2122/97 SLEKSDSLGLQIVRTLVSAELDGSLGMRDARERGTDVVLRVPVGRRGRLM
MMAR_1337|M.marinum_M SLEKSDSLGLQIVRTLVSAELDGSLGMREAPGRGTDVVLRVPIGRRGRLL
MUL_2542|M.ulcerans_Agy99 SLEKSDSLGLQIVRTLVSAELDGSLGMREAPGRGTDVVLRVPIGRRGRLL
MAV_4168|M.avium_104 SLEASDSLGLQIVRTLVSAELDGTLGMSEASERGTDVVLRVPIGRRTRML
MLBr_00803|M.leprae_Br4923 NLERSDSLGLQIVRTLVSSELDGSLGMRKAPNRGTDVVLRVPIGRRARLL
MAB_3540c|M.abscessus_ATCC_199 SVETSDRLGLQIVRTLVSAEIDGSLDMRSDPEGGTNAVLRVPLGRRARLM
.:* ** **********::*:**::.* **:. ****:*** *
Mflv_4686|M.gilvum_PYR-GCK QY
Mvan_1781|M.vanbaalenii_PYR-1 QY
MSMEG_1918|M.smegmatis_MC2_155 Q-
TH_0668|M.thermoresistible__bu Q-
Rv3220c|M.tuberculosis_H37Rv L-
Mb3246c|M.bovis_AF2122/97 L-
MMAR_1337|M.marinum_M A-
MUL_2542|M.ulcerans_Agy99 A-
MAV_4168|M.avium_104 L-
MLBr_00803|M.leprae_Br4923 L-
MAB_3540c|M.abscessus_ATCC_199 A-