For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YRVFEALDELSAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGELDDAQDVLDARDSMLQDAKTHAD SMVSSATTEAESILNHARTEADRILSDAKAQADRMVSEARQHSERMVADAREEAIRIATAAKREYEASVS RAQAECDRLIENGNISYEKAVQEGIKEQQRLVSQNEVVAAANAESTRLVDTAHAEADRLRGECDIYVDNK LAEFEEFLNGTLRSVGRGRHQLRTAAGTHDYAVR*
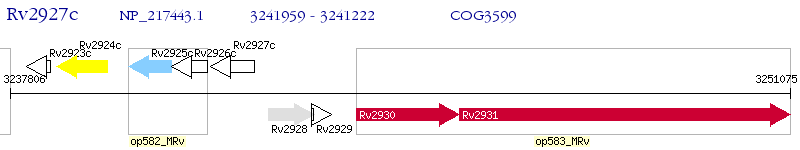
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2927c | - | - | 100% (245) | hypothetical protein Rv2927c |
| M. tuberculosis H37Rv | Rv2145c | wag31 | 6e-11 | 31.93% (119) | hypothetical protein Rv2145c |
| M. tuberculosis H37Rv | Rv1682 | - | 3e-06 | 23.88% (134) | coiled-coil structural protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2952c | - | 1e-134 | 100.00% (245) | hypothetical protein Mb2952c |
| M. gilvum PYR-GCK | Mflv_4194 | - | 1e-116 | 83.27% (245) | hypothetical protein Mflv_4194 |
| M. leprae Br4923 | MLBr_01661 | - | 1e-125 | 90.61% (245) | hypothetical protein MLBr_01661 |
| M. abscessus ATCC 19977 | MAB_3258c | - | 1e-107 | 77.14% (245) | hypothetical protein MAB_3258c |
| M. marinum M | MMAR_1780 | - | 1e-123 | 90.61% (245) | hypothetical protein MMAR_1780 |
| M. avium 104 | MAV_3785 | - | 1e-127 | 93.47% (245) | hypothetical protein MAV_3785 |
| M. smegmatis MC2 155 | MSMEG_2416 | - | 1e-117 | 84.08% (245) | hypothetical protein MSMEG_2416 |
| M. thermoresistible (build 8) | TH_1226 | - | 1e-102 | 80.27% (223) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2028 | - | 1e-123 | 90.20% (245) | hypothetical protein MUL_2028 |
| M. vanbaalenii PYR-1 | Mvan_2169 | - | 1e-116 | 84.08% (245) | hypothetical protein Mvan_2169 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3258c|M.abscessus_ATCC_199 --MYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
MSMEG_2416|M.smegmatis_MC2_155 --MYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
Mflv_4194|M.gilvum_PYR-GCK --MYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
Mvan_2169|M.vanbaalenii_PYR-1 --MYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
TH_1226|M.thermoresistible__bu ------------------------MTAGCVVPRGDVLELIDDIKDAIPGE
Rv2927c|M.tuberculosis_H37Rv --MYRVFEALDELSAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
Mb2952c|M.bovis_AF2122/97 --MYRVFEALDELSAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
MAV_3785|M.avium_104 --MYRVFEALDELSAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
MMAR_1780|M.marinum_M MAVYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELLDDIKDAIPGE
MUL_2028|M.ulcerans_Agy99 MAVYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELLDDIKDAIPGE
MLBr_01661|M.leprae_Br4923 MAVYRVFEALDELGAIVEEARGVPMTAGCVVPRGDVLELIDDIKDAIPGE
***************:**********
MAB_3258c|M.abscessus_ATCC_199 LDDAQDVLDARDSLLREAKEHHETVIGNSNAEAEQTLSHARNEADRLLAD
MSMEG_2416|M.smegmatis_MC2_155 LDDAQDVLDARDSLLREAKEHSESVISGANAEADSVLSHARAEADRLLAD
Mflv_4194|M.gilvum_PYR-GCK LDDAQDVLDARDAMLREAKDHAESTVSTANAEADSMVNHARTEADRLLAD
Mvan_2169|M.vanbaalenii_PYR-1 LDDAQDVLDARDSLLREAKDHADSTVSTANAEADSMVNHARAEADRLLAD
TH_1226|M.thermoresistible__bu LDDAQDVLDARDTLLREAKEHSESMVSTATAEAESLVSHARAEADRLLAE
Rv2927c|M.tuberculosis_H37Rv LDDAQDVLDARDSMLQDAKTHADSMVSSATTEAESILNHARTEADRILSD
Mb2952c|M.bovis_AF2122/97 LDDAQDVLDARDSMLQDAKTHADSMVSSATTEAESILNHARTEADRILSD
MAV_3785|M.avium_104 LDDAQDVLDARDSMLSDAKTHAESMVSSATTESESMLNHARAEADRVLSD
MMAR_1780|M.marinum_M LDDAQDVLDARDSMLQEAKAHADSMVSSATTESESMLNHARAEADRILSD
MUL_2028|M.ulcerans_Agy99 LDDAQDVLDARDSMLQEATAHADSMVSSATTESESMLNHARAEADRILSD
MLBr_01661|M.leprae_Br4923 LDDAQDVLDARDAMLNDAKAHADSMVSSATTESESLLSHARAEADRILSD
************::* :*. * :: :. :.:*::. :.*** ****:*::
MAB_3258c|M.abscessus_ATCC_199 AKAQADRMVAEARNHAEQTVGEAREEAARTIANAKREQESTIARAKAEAD
MSMEG_2416|M.smegmatis_MC2_155 AKAQADRMVAEARQHSERMVTEAREEAARLAAAAKREYEASTGRAKAEAD
Mflv_4194|M.gilvum_PYR-GCK AKAQADRMVAEARQHSERMVGEAREEAARIAATAKREYEASTGRAKSEAD
Mvan_2169|M.vanbaalenii_PYR-1 AKAQADRMVAEARQHSERMVAEARDEAARIAATAKREYEATTGRAKSEAD
TH_1226|M.thermoresistible__bu AKAQADRMVAEARQHSERMVTEAREEASRIVATAKREYEATTGRAKSEAD
Rv2927c|M.tuberculosis_H37Rv AKAQADRMVSEARQHSERMVADAREEAIRIATAAKREYEASVSRAQAECD
Mb2952c|M.bovis_AF2122/97 AKAQADRMVSEARQHSERMVADAREEAIRIATAAKREYEASVSRAQAECD
MAV_3785|M.avium_104 AKAQADRMVSEARQHSERMVAEAREEAVRIATAAKREYEASVSRAQAEAD
MMAR_1780|M.marinum_M AKAQADRMVGEARQHSERMVGEAREEAIRIAAAAKREYEASMSRAKSECD
MUL_2028|M.ulcerans_Agy99 AKAQADRMVGEARQHSERMVGEAREEAIRIAAAAKREYEASMSRAKSECD
MLBr_01661|M.leprae_Br4923 AKSQVDRMASEARQHSERMLGDAREESIRIATVAKREYEASLNRAQSECD
**:*.***..***:*:*: : :**:*: * : **** *:: **::*.*
MAB_3258c|M.abscessus_ATCC_199 RLVDSGNASYDKAVQEGIKEQQRLVAQTEVVQAAHAESTRLIDAAHAEAD
MSMEG_2416|M.smegmatis_MC2_155 RLIENGNISYEKAIQEGIKEQQRLVSQTEIVATANAEATRLIDAAHAEAD
Mflv_4194|M.gilvum_PYR-GCK RLLESGNLAYEKAVQEGIKEQQRLVSQTEVVATATAEATRMIDSAHAEAD
Mvan_2169|M.vanbaalenii_PYR-1 RLIENGNLTYEKAVQEGIKEQQRLVSQTEVVATATAEATRMIDSAHAEAD
TH_1226|M.thermoresistible__bu RLIESGNLAYEKAVQEGIKEQQRLVSQTEIVQTANAEASRLIDSAHAEAD
Rv2927c|M.tuberculosis_H37Rv RLIENGNISYEKAVQEGIKEQQRLVSQNEVVAAANAESTRLVDTAHAEAD
Mb2952c|M.bovis_AF2122/97 RLIENGNISYEKAVQEGIKEQQRLVSQNEVVAAANAESTRLVDTAHAEAD
MAV_3785|M.avium_104 RLLENGNISYEKAVQEGIKEQQRLVSQNSVVEAAHAEATRLIDSAHAEAD
MMAR_1780|M.marinum_M RLIESGNISYENAVQEGIKEQQRLVSQNEVVQAANAEATRLVDTAHAESD
MUL_2028|M.ulcerans_Agy99 RLIESGNISYENAVQEGIKEQQRLVSQNEVVQAANAEATRLVDTAHAESD
MLBr_01661|M.leprae_Br4923 RLIENGNISYEKAIQEGIKEQQRLVSQNEVVQAANAESTRLIDTAHAEAD
**::.** :*::*:***********:*..:* :* **::*::*:****:*
MAB_3258c|M.abscessus_ATCC_199 RLRGECDIYVDNKLAEFEDYLNGTLRSVGRGRHQLRTGAGTHDYVQR
MSMEG_2416|M.smegmatis_MC2_155 RLRGECDIYVDSKLAEFEEFLNGTLRSVGRGRHQLRTTAGTHDYVTR
Mflv_4194|M.gilvum_PYR-GCK RLRGECDIYVDSKLAEFEDFLNGTLRSVGRGRHQLRTSAGTHDYAAR
Mvan_2169|M.vanbaalenii_PYR-1 RLRGECDIYVDSKLAEFEDFLNGTLRSVGRGRHQLRTAAGTHDYAAR
TH_1226|M.thermoresistible__bu RLRGECDIYVDSKLAEFEEFLNGTLRSVNRGRHQLRTAAGTHDYVTR
Rv2927c|M.tuberculosis_H37Rv RLRGECDIYVDNKLAEFEEFLNGTLRSVGRGRHQLRTAAGTHDYAVR
Mb2952c|M.bovis_AF2122/97 RLRGECDIYVDNKLAEFEEFLNGTLRSVGRGRHQLRTAAGTHDYAVR
MAV_3785|M.avium_104 RLRGECDIYVDNKLAEFEEFLNGTLRSVGRGRHQLRTAAGTHDYAVR
MMAR_1780|M.marinum_M RLRGECDVYVDTKLAEFEEMLNGTLRSVGRGRHQLRTAAGTHDYATR
MUL_2028|M.ulcerans_Agy99 RLRGECDVYVDTKLAEFEEMLNGTLRSVGRGRHQLRTAAGTHDYATR
MLBr_01661|M.leprae_Br4923 RLRGECDIYVDNKLAEFEEFLNGTLRSVGRGRHQLRTAAGTHDYVTR
*******:***.******: ********.******** ******. *