For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRRGEIWQVDLDPARGSEANNQRPAVVVSNDRANATATRLGRGVITVVPVTSNIAKVYPFQVLLSATTTG LQVDCKAQAEQIRSIATERLLRPIGRVSAAELAQLDEALKLHLDLWS*
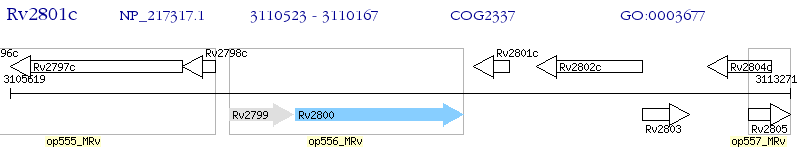
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2801c | - | - | 100% (118) | hypothetical protein Rv2801c |
| M. tuberculosis H37Rv | Rv1991c | - | 1e-09 | 35.04% (117) | hypothetical protein Rv1991c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2824c | - | 3e-62 | 100.00% (118) | hypothetical protein Mb2824c |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4448 | - | 1e-07 | 25.86% (116) | transcriptional modulator of MazE |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1144 | - | 2e-43 | 73.04% (115) | transcriptional modulator of MazE/toxin, MazF |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2801c|M.tuberculosis_H37Rv MMRRGEIWQVDLDPARGSEANNQRPAVVVSNDRANATATRLGRGVITVVP
Mb2824c|M.bovis_AF2122/97 MMRRGEIWQVDLDPARGSEANNQRPAVVVSNDRANATATRLGRGVITVVP
Mvan_1144|M.vanbaalenii_PYR-1 -MQRGDIWYVDLEPSRASEANKTRPAVIVSNDRANATASRLGRGVISVVP
MSMEG_4448|M.smegmatis_MC2_155 -MRRGDIYTA---AARGAYTGKPRPVLIIQDDRFDATAS------VTVVP
*:**:*: . .:*.: :.: **.:::.:** :***: ::***
Rv2801c|M.tuberculosis_H37Rv VTSNIAKVYPFQVLLSAT-TTGLQVDCKAQAEQIRSIATERLLRPIGRVS
Mb2824c|M.bovis_AF2122/97 VTSNIAKVYPFQVLLSAT-TTGLQVDCKAQAEQIRSIATERLLRPIGRVS
Mvan_1144|M.vanbaalenii_PYR-1 VTSNVDRVYPFQVLLQAP-DSGLAVASKAQAEQVRSIAAQRLIRRIGRVS
MSMEG_4448|M.smegmatis_MC2_155 FTTSDTDAPLLRIRIEPTPATGLSTVSSLMIDKVTTVPRSSLTHRVGRLA
.*:. . ::: :... :** . .. ::: ::. . * : :**::
Rv2801c|M.tuberculosis_H37Rv AAELAQLDEALKLHLDLWS
Mb2824c|M.bovis_AF2122/97 AAELAQLDEALKLHLDLWS
Mvan_1144|M.vanbaalenii_PYR-1 ARELAELDDALRLHLAL--
MSMEG_4448|M.smegmatis_MC2_155 ETDMVRTDRALLVFLGIAG
::.. * ** :.* :