For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRRGDIYTAAARGAYTGKPRPVLIIQDDRFDATASVTVVPFTTSDTDAPLLRIRIEPTPATGLSTVSSLM IDKVTTVPRSSLTHRVGRLAETDMVRTDRALLVFLGIAG
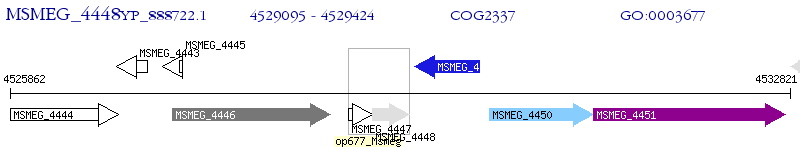
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4448 | - | - | 100% (109) | transcriptional modulator of MazE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2824c | - | 6e-08 | 25.86% (116) | hypothetical protein Mb2824c |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2801c | - | 6e-08 | 25.86% (116) | hypothetical protein Rv2801c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2930 | - | 6e-24 | 50.91% (110) | transcriptional modulator of MazE/toxin, MazF |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4448|M.smegmatis_MC2_155 -MRRGDIYTAAAR---GAYTGKPRPVLIIQDDRFDATAS------VTVVP
Mvan_2930|M.vanbaalenii_PYR-1 -MNRGEIWTVAG----GVYAAKPRPAVIVQDDLFDATSS------VTVAP
Mb2824c|M.bovis_AF2122/97 MMRRGEIWQVDLDPARGSEANNQRPAVVVSNDRANATATRLGRGVITVVP
Rv2801c|M.tuberculosis_H37Rv MMRRGEIWQVDLDPARGSEANNQRPAVVVSNDRANATATRLGRGVITVVP
*.**:*: . * : : **.:::.:* :**:: :**.*
MSMEG_4448|M.smegmatis_MC2_155 FTTSDTDAPLLRIRIEPTP--ATGLSTVSSLMIDKVTTVPRSSLTHRVGR
Mvan_2930|M.vanbaalenii_PYR-1 MTSTLLDAPLMRIRIAGGDGRLSGLDHDRDVMIDKLTTVRRSNVHVRVGR
Mb2824c|M.bovis_AF2122/97 VTSNIAKVYPFQVLLSATT---TGLQVDCKAQAEQIRSIATERLLRPIGR
Rv2801c|M.tuberculosis_H37Rv VTSNIAKVYPFQVLLSATT---TGLQVDCKAQAEQIRSIATERLLRPIGR
.*:. .. ::: : :**. . ::: :: . : :**
MSMEG_4448|M.smegmatis_MC2_155 LAETDMVRTDRALLVFLGIAG
Mvan_2930|M.vanbaalenii_PYR-1 LTAEQVVEVERTMMAFLGLAR
Mb2824c|M.bovis_AF2122/97 VSAAELAQLDEALKLHLDLWS
Rv2801c|M.tuberculosis_H37Rv VSAAELAQLDEALKLHLDLWS
:: ::.. :.:: .*.: