For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRADEEPGDLSAVAQDYLKVIWTAQEWSQDKVSTKMLAERIGVSASTASESIRKLAEQGLVDHEKYGAVT LTDSGRRAALAMVRRHRLLETFLVNELGYRWDEVHDEAEVLEHAVSDRLMARIDAKLGFPQRDPHGDPIP GADGQVPTPPARQLWACRDGDTGTVARISDADPQMLRYFASIGISLDSRLRVLARREFAGMISVAIDSAD GATVDLGSPAAQAIWVVS
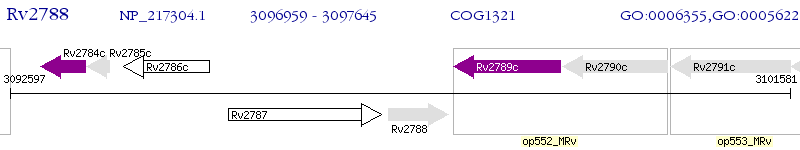
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2788 | sirR | - | 100% (228) | PROBABLE TRANSCRIPTIONAL REPRESSOR SIRR |
| M. tuberculosis H37Rv | Rv2711 | ideR | 5e-17 | 30.65% (186) | IRON-dependent repressor and activator IDER |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2811 | sirR | 1e-128 | 99.56% (228) | transcriptional repressor SIRR |
| M. gilvum PYR-GCK | Mflv_4026 | - | 1e-105 | 80.69% (233) | iron dependent repressor |
| M. leprae Br4923 | MLBr_01013 | - | 2e-17 | 34.59% (133) | iron dependent repressor |
| M. abscessus ATCC 19977 | MAB_3110 | - | 4e-98 | 75.64% (234) | iron dependent transcriptional repressor FeoA |
| M. marinum M | MMAR_1920 | sirR | 1e-116 | 89.57% (230) | transcriptional repressor SirR |
| M. avium 104 | MAV_3679 | - | 1e-110 | 85.04% (234) | iron repressor protein |
| M. smegmatis MC2 155 | MSMEG_2652 | - | 1e-104 | 80.69% (233) | iron repressor protein |
| M. thermoresistible (build 8) | TH_1279 | sirR | 2e-89 | 79.31% (203) | PROBABLE TRANSCRIPTIONAL REPRESSOR SIRR |
| M. ulcerans Agy99 | MUL_2145 | sirR | 1e-116 | 88.70% (230) | transcriptional repressor SirR |
| M. vanbaalenii PYR-1 | Mvan_2347 | - | 1e-104 | 78.66% (239) | iron dependent repressor |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2788|M.tuberculosis_H37Rv MRADEE-------PGDLSAVAQDYLKVIWTAQEWSQD----KVSTKMLAE
Mb2811|M.bovis_AF2122/97 MRADEE-------PGDLSAVAQDYLKVIWTAQEWSQD----KVSTKMLAE
MMAR_1920|M.marinum_M MRADEE-------PGGLTTVAQDYLKVIWTAQEWSRE----RVSTKMLAE
MUL_2145|M.ulcerans_Agy99 MRADEE-------PGGLTTVAQDYLKVIWTAQEWSRE----RVSTKMLAE
MAV_3679|M.avium_104 MRADEE-------RGGLTAVGQDYLKAIWNAQEWSPEGAPQKVSTKMLAE
Mflv_4026|M.gilvum_PYR-GCK MSPDGNP----ANPTDLSTVAQDYLKVIWTAQEWSHE----KVSTKLLAD
Mvan_2347|M.vanbaalenii_PYR-1 MSPDGNP----ANPTDLSTVAQDYLKVIWTAQEWSRE----KVSTKLLAE
MSMEG_2652|M.smegmatis_MC2_155 MSPDGD-------HRDLSSVAQDYLKVIWTAQEWSRE----KVSTKLLAE
TH_1279|M.thermoresistible__bu -----------------------------------LE----KVSTKLLAE
MAB_3110|M.abscessus_ATCC_1997 MSPTRSDRPAGSADTDLTTVAQDYLKVIWTAQEWSHE----KVSTKMLAE
MLBr_01013|M.leprae_Br4923 -------------MNDLVDTTEMYLRTIYDLEEEGVT-----PLRARIAE
:*:
Rv2788|M.tuberculosis_H37Rv RIGVSASTASESIRKLAEQGLVDHEKYGAVTLTDSGRRAALAMVRRHRLL
Mb2811|M.bovis_AF2122/97 RIGVSASTASESIRKLAEQGLVDHEKYGAVTLTDSGRRAALAMVRRHRLL
MMAR_1920|M.marinum_M KIGVSASTASESIRKLAEQGLVDHEKYGAVTLTDSGRRAALAMVRRHRLL
MUL_2145|M.ulcerans_Agy99 KIGVSASTASESIRKLAEQGLVDHEKYGAVTLTDSGRRAALAMVRRHRLL
MAV_3679|M.avium_104 KIGVSASTASESIRKLAEQGLVDHEKYGAVTLTEAGRRAALEMVRRHRLL
Mflv_4026|M.gilvum_PYR-GCK RIGVSASTASESIRKLADQGLVDHEKYGAVTLTDAGRRAALAMVRRHRLM
Mvan_2347|M.vanbaalenii_PYR-1 RIGVSASTASESIRKLADQGLVDHEKYGAVTLTEAGRRAALAMVRRHRLM
MSMEG_2652|M.smegmatis_MC2_155 RIGVSASTASESIRKLADQGLVHHEKYGAVTLTDAGRRAALAMVRRHRLM
TH_1279|M.thermoresistible__bu RLGVSPSTASEAIRRLADQGLVDHEKYGAVTLTEAGRRAALAMVRRHRLL
MAB_3110|M.abscessus_ATCC_1997 RMGVSASTASESIRKLADQGLVDHEKYGAVTLTDHGRKAAVLMVRRHRLL
MLBr_01013|M.leprae_Br4923 RLEQSGPTVSQTVSRMERDGLLRVAGNRHLELTTKGRAMAIAVMRKHRLA
:: * .*.*::: :: :**: : ** ** *: ::*:***
Rv2788|M.tuberculosis_H37Rv ETFLVNELGYRWDEVHDEAEVLEHAVSDRLMARIDAKLGFPQRDPHGDPI
Mb2811|M.bovis_AF2122/97 ETFLVNELGYRWDEVHDEAEVLEHAVSDRLMARIDAKLGFPQRDPHGAPI
MMAR_1920|M.marinum_M ETFLVNELGYGWDEVHDEAEVLEHAVSDRLVARIDAKLGFPRRDPHGDPI
MUL_2145|M.ulcerans_Agy99 ETFLVNELGYGWDEVHDEAEVLEHAVSDRLVTRIDAKLGFPRRDPHGDPI
MAV_3679|M.avium_104 ETFLVNELGYAWDEVHDEAEVLEHAVSDRLVARIDAKLGFPQRDPHGDPI
Mflv_4026|M.gilvum_PYR-GCK ETFLVNELGYGWDEVHDEAEILEHAVSDRMLDRIDAKLGYPTRDPHGDPI
Mvan_2347|M.vanbaalenii_PYR-1 ETFLVNELGYSWDEVHDEAEVLEHAVSDRMLDRIDAKLGYPTRDPHGDPI
MSMEG_2652|M.smegmatis_MC2_155 ETFLVQELGYGWDEVHDEAEILEHAVSDRMLDRIDAKLGHPTRDPHGDPI
TH_1279|M.thermoresistible__bu ETFLVQELGYSWDEVHDEAEVLEHAVSDLMLDRIDAKLGYPTRDPHGDPI
MAB_3110|M.abscessus_ATCC_1997 ETYLVNELGYGWDEVHDEAEVLEHAVSDLLLAKIDAKLGHPQRDPHGDPI
MLBr_01013|M.leprae_Br4923 ERLLVDVIGLPWEEVHAEACRWEHVMSEDVERRLIKVLNNPTTSPFGNPI
* **: :* *:*** ** **.:*: : :: *. * .*.* **
Rv2788|M.tuberculosis_H37Rv P-------GADGQVPTPPARQLWACRDGD-----TGTVARISDADPQMLR
Mb2811|M.bovis_AF2122/97 P-------GADGQVPTPPARQLWACRDGD-----TGTVARISDADPQMLR
MMAR_1920|M.marinum_M P-------ASDGQVPTPPARQLWACRDGD-----TATVARISDADPEMLR
MUL_2145|M.ulcerans_Agy99 P-------SSDGQVPTPPARQLWACRDGD-----TATVARISDADPEILR
MAV_3679|M.avium_104 P-------ATDGQVPTPPARQLWACGDGD-----TGTVARISDADSEMLR
Mflv_4026|M.gilvum_PYR-GCK P-------AADGKVPTPPARQLSACQDGE-----AGTVARISDADPEMLR
Mvan_2347|M.vanbaalenii_PYR-1 P-------AADGRVPTPPARQLSDCQDGE-----AGTVARISDADPEMLR
MSMEG_2652|M.smegmatis_MC2_155 P-------AADGQVPTPPARQLSVCQDGD-----AGTVARISDADPEMLR
TH_1279|M.thermoresistible__bu P-------RADGQVPTPPARQLSACPEGD-----VGMVARISDADPEMLR
MAB_3110|M.abscessus_ATCC_1997 P-------GPDGQVPTPPARQLSDCANGD-----TGVVARISDSDPEMLR
MLBr_01013|M.leprae_Br4923 PGLLDLGAGPDASAANAKLVRLTELPSGSPVAVVVRQLTEHVQDDIDLIT
* .*. .... :* .*. . ::. : * :::
Rv2788|M.tuberculosis_H37Rv YFASIGISLDSRLRVLARREFAGMISVAIDSA---------------DGA
Mb2811|M.bovis_AF2122/97 YFASIGISLDSRLRVLARREFAGMISVAIDSA---------------DGA
MMAR_1920|M.marinum_M YFDSVGINLDSQLRVLTRREFAGMISVAIESA---------------DGA
MUL_2145|M.ulcerans_Agy99 YFDSVGINLDSQLRVLTRREFAGMISVAIESA---------------DGA
MAV_3679|M.avium_104 YFTDVGINLDSRLRVLTRREFAGMISVAIESG---------------DGA
Mflv_4026|M.gilvum_PYR-GCK YFDTVGISLDSRLRVVARRDFAGMISVAVENP------GSSGGSKAASDG
Mvan_2347|M.vanbaalenii_PYR-1 YFDAVGINLDSRLRVVARRDFAGMISVAIENPERPENPSNPDGATSGHDN
MSMEG_2652|M.smegmatis_MC2_155 YFDSVGISLDSRLRVVARRDFAGTISVAIEDP-----------AVPEGDP
TH_1279|M.thermoresistible__bu YFDTVGISLDSRLRVVARRDFAGMISVAVTAP-------------DRGEP
MAB_3110|M.abscessus_ATCC_1997 YFDTVGVALDARVTVDERRDFAGTISVSIDSD-------------EPGRD
MLBr_01013|M.leprae_Br4923 RLKDTGVVPNARVTVETSP--AGNVIIIIPGH------------------
: *: :::: * ** : : :
Rv2788|M.tuberculosis_H37Rv T--VDLGSPAAQAIWVVS--
Mb2811|M.bovis_AF2122/97 T--VDLGSPAAQAIWVVS--
MMAR_1920|M.marinum_M ANTVDLGSPAARAIWVVA--
MUL_2145|M.ulcerans_Agy99 ANTVDLGSPAARAIWVVA--
MAV_3679|M.avium_104 ETTVDLGSPAARAIWVTA--
Mflv_4026|M.gilvum_PYR-GCK ETTVDLGSPAAQAIWVVA--
Mvan_2347|M.vanbaalenii_PYR-1 ETTVDLGSPAAQAIWVTA--
MSMEG_2652|M.smegmatis_MC2_155 EATVDLGSPAAEAIWVVGAS
TH_1279|M.thermoresistible__bu DVTVDLGHPAAEAIWVTSAG
MAB_3110|M.abscessus_ATCC_1997 ERKLELGNPAAQSIWVVLD-
MLBr_01013|M.leprae_Br4923 -ENVTLPHEMAHAVKVEKV-
: * *.:: *