For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SFLTTQPEELAAAAGKLETIGSAMVAQNAAAAAPTTTGVIPAAADEISVLQAPLFTAYGTLYQQVSAEAA AVYDLFVKTLGVSAGTYAATEAANSSAAASPLSGIASILGSTPGKVPSWISDIANIFNIGAGNWASAASD LLGLASGGLLPAAEEAALEEGLEGAGLSELGAAEAAVGEAPIAAGLGAAPLAAGLSRASSIGALSVPPSW AGQANLVSSTSTLQGAGWTTAAPHGAAGTVIPGMPGLASATRSSAGFGAPRYGAKPIVVPKPAV*
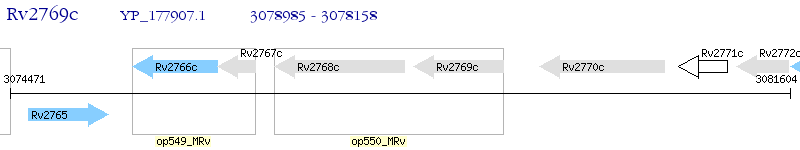
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2769c | PE27 | - | 100% (275) | PE FAMILY PROTEIN |
| M. tuberculosis H37Rv | Rv1040c | PE8 | 1e-94 | 67.14% (283) | PE family protein |
| M. tuberculosis H37Rv | Rv1788 | PE18 | 1e-30 | 66.67% (99) | PE family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2791c | PE27 | 1e-149 | 99.27% (275) | PE family protein |
| M. gilvum PYR-GCK | Mflv_0326 | - | 1e-06 | 34.44% (90) | PE domain-containing protein |
| M. leprae Br4923 | MLBr_01183 | - | 2e-14 | 41.94% (93) | hypothetical protein MLBr_01183 |
| M. abscessus ATCC 19977 | MAB_0664 | - | 8e-09 | 37.50% (104) | PE family protein |
| M. marinum M | MMAR_1949 | - | 1e-100 | 71.48% (277) | PE family protein |
| M. avium 104 | MAV_1346 | - | 1e-53 | 47.54% (284) | PE family protein |
| M. smegmatis MC2 155 | MSMEG_0618 | - | 1e-05 | 34.65% (101) | PE family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4620 | - | 7e-86 | 62.19% (283) | PE family protein |
| M. vanbaalenii PYR-1 | Mvan_0414 | - | 8e-08 | 35.79% (95) | PE domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2769c|M.tuberculosis_H37Rv MS-FLTTQPEELAAAAGKLETIGSAMVAQNAAAAAPTTTGVIPAAADEIS
Mb2791c|M.bovis_AF2122/97 MS-FLTTQPEELAAAAGKLETIGSAMVAQNAAAAAPTTTGVIPAAADEIS
MMAR_1949|M.marinum_M MS-FLTTQPEELAAAAGKLETIGATMDAENVAAVAPTTTGVIPAAADEVS
MUL_4620|M.ulcerans_Agy99 MS-FLKTVPEELTAAAAQLGTIGSSMSAQNAAAAAPTTV-IAPAALDEVS
MLBr_01183|M.leprae_Br4923 MPLFLNAEPQALTAAANTLEGLSAATVASNAAAAQLTTE-IAPPAADDVS
MAV_1346|M.avium_104 MS-FVTTRPEALLAVASMLEGLGSSMDAQNAAAAAPTTS-IAPAAADEVS
Mflv_0326|M.gilvum_PYR-GCK MT--LRVVPEGLTAASAAVEALTARMAAAHAAAAPLVTA-VLPPAADAVS
Mvan_0414|M.vanbaalenii_PYR-1 MT--LRVVPEGLTAAGAAVEALTARMAAAHAAAAPVVTA-VLPPAADVVS
MSMEG_0618|M.smegmatis_MC2_155 MT--LRVVPEGLTAASSAVEALTARLAAAHAAAAPMIST-VLPPAADAVS
MAB_0664|M.abscessus_ATCC_1997 MS--LNVVPEGLTAASAAVEALTARLAAVNAAAAPVIGA-VMPPAADPVS
*. : . *: * *.. : : : * :.**. : *.* * :*
Rv2769c|M.tuberculosis_H37Rv VLQAPLFTAYGTLYQQVSAEAAAVYDLFVKTLGVSAGTYAATEAAN-SSA
Mb2791c|M.bovis_AF2122/97 VLQASLFTAYGTLYQQVSAEAAAVYDLFVKTLGVSAGTYAATEAAN-SSA
MMAR_1949|M.marinum_M ALQASLFTAYGTLYQQVSAEAATLYDMFVRTLGVSAGTYAAAETAN-SSA
MUL_4620|M.ulcerans_Agy99 ALQAALFTAYGTYYQQVSADAQAMHDMFVNTLGASAGTYGATENLN-SAA
MLBr_01183|M.leprae_Br4923 ILLAHFFSGHGRQYQAHASQGATNHQDLIQSLLTSSSAYAGTETAN----
MAV_1346|M.avium_104 ALQAAQFSAYGTWYQQVSAQAKAIHQTLVQNLDTNADSYGATEAANRVNT
Mflv_0326|M.gilvum_PYR-GCK LQTATGLSANGVEHQSIAARGVEELGRSGVGVGDSAASYVSGDAMA----
Mvan_0414|M.vanbaalenii_PYR-1 LQAATGFSARAAEHQSVAAQGVEELGRSGIAVGESAASYAGGDAMA----
MSMEG_0618|M.smegmatis_MC2_155 LQTAAGFSANGAQQSAVAAQGVEELGRSGVGVGESGVSYATGDAQA----
MAB_0664|M.abscessus_ATCC_1997 MQSAALFSAHGLERTGAGARAAYELGRSGVGATEAAASYTVGDIQA----
* ::. . .: . . :* :
Rv2769c|M.tuberculosis_H37Rv AASPLSGIAS----ILGSTPGKVPS-WISDIANIFNIGAGNWASAASDLL
Mb2791c|M.bovis_AF2122/97 AASPLSGIAS----ILGSTPGKVPS-WISDIANIFNIGAGNWASAASDLL
MMAR_1949|M.marinum_M AASPLSGIID----ALGAS---TP---ASDIANIVNIGAGNWASASSDLL
MUL_4620|M.ulcerans_Agy99 AASPLDGITSGISGFIQNTGSIIPDSWSNNIGNLTNIGVGNWASASSTLI
MLBr_01183|M.leprae_Br4923 ----------------------------HDSL------------------
MAV_1346|M.avium_104 SAAALQGLAPNV--IPNAAPAADPPPAGTTSLGTSIGWAQNVGAAASDFI
Mflv_0326|M.gilvum_PYR-GCK --------------------------------------------ATSYLI
Mvan_0414|M.vanbaalenii_PYR-1 --------------------------------------------ASSYLI
MSMEG_0618|M.smegmatis_MC2_155 --------------------------------------------AASYLT
MAB_0664|M.abscessus_ATCC_1997 --------------------------------------------AATYLP
Rv2769c|M.tuberculosis_H37Rv GLASGGLLPAAEEAALEEGLEGAGLSELGAAEAAVGEAPIAAGLGAAPLA
Mb2791c|M.bovis_AF2122/97 GLASGGLLPAAEEAALEEGLEGAGLSELGAAEAAVGEAPIAAGLGAAPLA
MMAR_1949|M.marinum_M ALAGGGLLPAAVQAAEGDVAQAAGSAESAAAAAG------AASLGEAPIV
MUL_4620|M.ulcerans_Agy99 GLAGGGLLP-AEEAAEADVSLGAEAGLGALSELG----GAEAALG-APIA
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 TLGEGQFAPVPGAAGIPNFNPGLTAATTAGPPAP----AAPAAAAGAPVL
Mflv_0326|M.gilvum_PYR-GCK ARG-----------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 ARG-----------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 ARGL----------------------------------------------
MAB_0664|M.abscessus_ATCC_1997 GIA-----------------------------------------------
Rv2769c|M.tuberculosis_H37Rv AGLSRASSIGALSVPPSWAG-QANLVSSTST--LQGAGWTTAAPHGAAGT
Mb2791c|M.bovis_AF2122/97 AGLSRASSIGALSVPPSWAG-QANLVSSTST--LQGAGWTTAAPHGAAGT
MMAR_1949|M.marinum_M AGVGGVSAMSAASASPSWAG-QATLVSSTSAGALRGAGWTVPAPQAAPGT
MUL_4620|M.ulcerans_Agy99 AGIGGASAVGMLSVPPTWAG-SATLVSSTTP--LAGAGWTAAAPTAAPGT
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 ASMGRGPSIGGLSVPPSWAPGGVAPAANATPAQLVGAGFTSAAPDTAPVT
Mflv_0326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0664|M.abscessus_ATCC_1997 --------------------------------------------------
Rv2769c|M.tuberculosis_H37Rv VIP-GMPGLASATRSSAGFGAPRYGAKPIVVPKPAV-
Mb2791c|M.bovis_AF2122/97 VIP-GMPGLASATRSSAGFGAPRYGAKPIVMPKPAV-
MMAR_1949|M.marinum_M VIP-GMPGLAAATRNSAGFGAPRYGVKPIVMPKPAAV
MUL_4620|M.ulcerans_Agy99 FMG-VPAGLGSAARNSAGFGAPRYGVKPIVMPKPAAV
MLBr_01183|M.leprae_Br4923 -------------------------------------
MAV_1346|M.avium_104 AVPGGVPSMATAGRGGYGLGAPRYGVKPTVMPKPAI-
Mflv_0326|M.gilvum_PYR-GCK -------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 -------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 -------------------------------------
MAB_0664|M.abscessus_ATCC_1997 -------------------------------------