For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LQQIRGPADLQHLSQAQLRELAAEIREFLIHKVAATGGHLGPNLGVVELTLALHRVFDSPHDPIIFDTGH QAYVHKMLTGRSQDFATLRKKGGLSGYPSRAESEHDWVESSHASAALSYADGLAKAFELTGHRNRHVVAV VGDGALTGGMCWEALNNIAASRRPVIIVVNDNGRSYAPTIGGVADHLATLRLQPAYEQALETGRDLVRAV PLVGGLWFRFLHSVKAGIKDSLSPQLLFTDLGLKYVGPVDGHDERAVEVALRSARRFGAPVIVHVVTRKG MGYPPAEADQAEQMHSTVPIDPATGQATKVAGPGWTATFSDALIGYAQKRRDIVAITAAMPGPTGLTAFG QRFPDRLFDVGIAEQHAMTSAAGLAMGGLHPVVAIYSTFLNRAFDQIMMDVALHKLPVTMVLDRAGITGS DGASHNGMWDLSMLGIVPGIRVAAPRDATRLREELGEALDVDDGPTALRFPKGDVGEDISALERRGGVDV LAAPADGLNHDVLLVAIGAFAPMALAVAKRLHNQGIGVTVIDPRWVLPVSDGVRELAVQHKLLVTLEDNG VNGGAGSAVSAALRRAEIDVPCRDVGLPQEFYEHASRSEVLADLGLTDQDVARRITGWVAALGTGVCASD AIPEHLD*
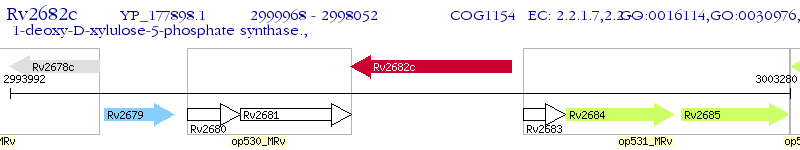
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2682c | dxs1 | - | 100% (638) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. tuberculosis H37Rv | Rv3379c | dxs2 | e-148 | 48.57% (558) | 1-deoxy-D-xylulose-5-phosphate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2701c | dxs1 | 0.0 | 100.00% (638) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. gilvum PYR-GCK | Mflv_3923 | - | 0.0 | 80.13% (624) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. leprae Br4923 | MLBr_01038 | dxs | 0.0 | 86.39% (632) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. abscessus ATCC 19977 | MAB_2990c | - | 0.0 | 75.91% (635) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. marinum M | MMAR_2032 | dxs1 | 0.0 | 90.30% (639) | 1-deoxy-D-xylulose 5-phosphate synthase Dxs1 |
| M. avium 104 | MAV_3577 | dxs | 0.0 | 87.54% (642) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. smegmatis MC2 155 | MSMEG_2776 | dxs | 0.0 | 79.47% (638) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. thermoresistible (build 8) | TH_2209 | dxs | 0.0 | 79.03% (639) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. ulcerans Agy99 | MUL_3319 | dxs1 | 0.0 | 89.51% (639) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. vanbaalenii PYR-1 | Mvan_2477 | - | 0.0 | 79.00% (638) | 1-deoxy-D-xylulose-5-phosphate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3923|M.gilvum_PYR-GCK MLEQIRGPADLQHLSRAQMEDLAREIRDFLIHKVAATGGHLGPNLGVVEL
Mvan_2477|M.vanbaalenii_PYR-1 MLEQIRGPADLQHLSQAQLEDLAHEIRDFLIHKVAATGGHLGPNLGVVEL
MSMEG_2776|M.smegmatis_MC2_155 MLEQVRGPADLQHLSQSQLRDLAQEIRQFLIHKVAATGGHLGPNLGVVEL
TH_2209|M.thermoresistible__bu MLEQIRGPADLQHLSKAQLRDLADEIRQFLIHKVAATGGHLGPNLGVVEL
Rv2682c|M.tuberculosis_H37Rv MLQQIRGPADLQHLSQAQLRELAAEIREFLIHKVAATGGHLGPNLGVVEL
Mb2701c|M.bovis_AF2122/97 MLQQIRGPADLQHLSQAQLRELAAEIREFLIHKVAATGGHLGPNLGVVEL
MMAR_2032|M.marinum_M MLQQIRGPADLQHLSQAQLRELAQEIREFLVHKVAATGGHLGPNLGVVEL
MUL_3319|M.ulcerans_Agy99 MLQQIRGPADLQHLSQAQLRELAQEIREFLVHKVAATGGHLGPNLGVVEL
MAV_3577|M.avium_104 MLEQIRGPADLQHLSTHQLRELAAEIREFLIHKVAATGGHLGPNLGVVEL
MLBr_01038|M.leprae_Br4923 MLEQIRRPADLQHLSQQQLRDLAAEIRELLVHKVAATGGHLGPNLGVVEL
MAB_2990c|M.abscessus_ATCC_199 MLDDIRGPGDLQGLSKRELDLLADEIREFLIHKVAATGGHLGPNLGVVEL
**:::* *.*** ** :: ** ***::*:*******************
Mflv_3923|M.gilvum_PYR-GCK TLALHRVFDSPHDPILFDTGHQAYVHKMLTGRSRDFDTLRKKDGLSGYPS
Mvan_2477|M.vanbaalenii_PYR-1 TLALHRVFDSPHDPILFDTGHQAYVHKMLTGRCRDFDSLRKKDGLSGYPS
MSMEG_2776|M.smegmatis_MC2_155 TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRSEDFDTLRSKDGLSGYPS
TH_2209|M.thermoresistible__bu TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRAADFDTLRKKGGLSGYPS
Rv2682c|M.tuberculosis_H37Rv TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRSQDFATLRKKGGLSGYPS
Mb2701c|M.bovis_AF2122/97 TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRSQDFATLRKKGGLSGYPS
MMAR_2032|M.marinum_M TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRAQDFESLRKKGGLSGYPC
MUL_3319|M.ulcerans_Agy99 TLALHRVFDSPHDPIIFDTGHQTYVHKMLTGRAQDFESLRKKGGLSGYPC
MAV_3577|M.avium_104 TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRAHEFESLRKKGGLSGYPS
MLBr_01038|M.leprae_Br4923 TLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRCQDFDSLRKKAGLSGYPS
MAB_2990c|M.abscessus_ATCC_199 SLALHRVFESPYDPIIFDTGHQAYVHKILTGRAGDFDTLRQKDGLSGYPS
:*******:**:***:******:****:****. :* :**.* ******.
Mflv_3923|M.gilvum_PYR-GCK RSESEHDWVESSHASSALSYADGLAKAFELNGHRNRHVVAVVGDGALTGG
Mvan_2477|M.vanbaalenii_PYR-1 SAESEHDWIESSHASSALSYADGLAKAFELSGHRNRHVVAVVGDGALTGG
MSMEG_2776|M.smegmatis_MC2_155 RSESDHDWVESSHASAALSYADGLAKAFELTGHRNRHVVAVVGDGALTGG
TH_2209|M.thermoresistible__bu RAESEHDWVESSHASAALSYADGLAKAFELTGHRNRHVVAVVGDGALTGG
Rv2682c|M.tuberculosis_H37Rv RAESEHDWVESSHASAALSYADGLAKAFELTGHRNRHVVAVVGDGALTGG
Mb2701c|M.bovis_AF2122/97 RAESEHDWVESSHASAALSYADGLAKAFELTGHRNRHVVAVVGDGALTGG
MMAR_2032|M.marinum_M RAESEHDWVESSHASAALSYADGLSKAFELSGLRNRHVVAVVGDGALTGG
MUL_3319|M.ulcerans_Agy99 RAESEHDWVESSHASAALSYADGLSKAFELSGLRNRHVVAVVGDGALTGG
MAV_3577|M.avium_104 RSESEHDWVESSHASAALSYADGLAKAFELSGHRNRHVVAVVGDGALTGG
MLBr_01038|M.leprae_Br4923 RAESEHDWVESSHASTALSYADGLAKAFELAGNRNRHVVAVVGDGALTGG
MAB_2990c|M.abscessus_ATCC_199 RAESEHDWVESSHASTALSYADGLAKAFELTGQRRRHVVAVVGDGAMTGG
:**:***:******:********:***** * *.***********:***
Mflv_3923|M.gilvum_PYR-GCK MCWEALNNIAASR-RPVVIVVNDNGRSYAPTIGGFAEHLAGLRLQPGYER
Mvan_2477|M.vanbaalenii_PYR-1 MCWEALNNIAASR-RPVVIVVNDNGRSYAPTIGGFAEHLAGLRLQPGYER
MSMEG_2776|M.smegmatis_MC2_155 MCWEALNNIATSR-RPVVIVVNDNGRSYAPTIGGLAEHLAALRLQPGYER
TH_2209|M.thermoresistible__bu MCWEAINNIAAAR-RPVVIVVNDNGRSYAPTIGGFAEHLAALRLQPQYEN
Rv2682c|M.tuberculosis_H37Rv MCWEALNNIAASR-RPVIIVVNDNGRSYAPTIGGVADHLATLRLQPAYEQ
Mb2701c|M.bovis_AF2122/97 MCWEALNNIAASR-RPVIIVVNDNGRSYAPTIGGVADHLATLRLQPAYEQ
MMAR_2032|M.marinum_M MCWEALNNIAASH-RPVVIVVNDNGRSYAPTIGGVADHLAKLRLQPAYEQ
MUL_3319|M.ulcerans_Agy99 MCWEALNNIAASH-RPVVIVVNDNGRSYAPTIGGVADHLAKLRLQPAYEQ
MAV_3577|M.avium_104 MCWEALNNIAASG-RPVIIVVNDNGRSYAPTIGGVADHLATLRLQPAYEQ
MLBr_01038|M.leprae_Br4923 MCWEALNNIAATP-RPVVIVVNDNGRSYAPTIGGVADHLATLRLQPAYER
MAB_2990c|M.abscessus_ATCC_199 MCWEALNNIAAAEHRRIIIVVNDNGRSYAPTIGGLASHLAGLRLTPSYEK
*****:****:: * ::****************.*.*** *** * **.
Mflv_3923|M.gilvum_PYR-GCK VLEEGRKAVRGVPMIGEFCYQCMHSIKAGIKDALSPQVMFTDLGLKYVGP
Mvan_2477|M.vanbaalenii_PYR-1 VLEEGRKAVRGVPMIGEFCYQCMHSIKVGIKDALSPQVMFTDLGLKYVGP
MSMEG_2776|M.smegmatis_MC2_155 VLEEGRKAVRGVPLIGELCYQCIHSIKAGIKDAIAPQVMFTDLGIKYVGP
TH_2209|M.thermoresistible__bu LLEQSRQAVRGIPMIGELCYRAMHSVKVGLKDALAPQEMFTDLGLKYVGP
Rv2682c|M.tuberculosis_H37Rv ALETGRDLVRAVPLVGGLWFRFLHSVKAGIKDSLSPQLLFTDLGLKYVGP
Mb2701c|M.bovis_AF2122/97 ALETGRDLVRAVPLVGGLWFRFLHSVKAGIKDSLSPQLLFTDLGLKYVGP
MMAR_2032|M.marinum_M VLEKGRDVVRAVPLVGEVCYHFMHSVKAGIKDSLSPQLLFTDLGLKYVGP
MUL_3319|M.ulcerans_Agy99 VLEKGRDVVRAVPLVGEVCYHFMHSVKAGIKDSLSPQLLFTDLGLKYVGP
MAV_3577|M.avium_104 ALQRGRDALRALPLVGKFAYRVMHSVKAGIKDSLSPQLLFTDLGLKYVGP
MLBr_01038|M.leprae_Br4923 LLEKGRDALHSLPLIGQIAYRFMHSVKAGIKDSLSPQLLFTDLGLKYVGP
MAB_2990c|M.abscessus_ATCC_199 VLDESKKLLRGMPVVGPLAYALMHSLKVGVKDAVSPQVMFTDLGLKYVGP
*: .:. ::.:*::* . : :**:*.*:**:::** :*****:*****
Mflv_3923|M.gilvum_PYR-GCK IDGHDESAVEGALRHARAFNAPVVVHVVTRKGMGYAPAENDVDEQMHACG
Mvan_2477|M.vanbaalenii_PYR-1 IDGHDEHAVESALRHARAFNAPVVVHVVTRKGMGYAPAENDADDQMHACG
MSMEG_2776|M.smegmatis_MC2_155 IDGHDEHAVESALRHARGFNAPVIVHVVTRKGMGYAPAENDEADQMHACG
TH_2209|M.thermoresistible__bu IDGHDEHAVEVALRRARGYNAPVVVHVVTRKGMGYGPAEADEAEQMHACG
Rv2682c|M.tuberculosis_H37Rv VDGHDERAVEVALRSARRFGAPVIVHVVTRKGMGYPPAEADQAEQMHSTV
Mb2701c|M.bovis_AF2122/97 VDGHDERAVEVALRSARRFGAPVIVHVVTRKGMGYPPAEADQAEQMHSTV
MMAR_2032|M.marinum_M VDGHDERAVEVALRSARGFGGPVIVHVVTRKGMGYGPAEADVAEQMHSTV
MUL_3319|M.ulcerans_Agy99 VDGHDERAVEVALRSARGFGGPVIVHVVTRKGMGYGPAEADVAEQMHSTV
MAV_3577|M.avium_104 VDGHDERAVEAALRHARGFGRPVIVHVVTRKGMGYAPAEDDEADQMHSCG
MLBr_01038|M.leprae_Br4923 VDGHDEHAVEVALRKARGFGGPVIVHVVTRKGMGYPPAEADQAEQMHTCG
MAB_2990c|M.abscessus_ATCC_199 VDGHDEHAVEDALRRARGYGGPVIVHVITRKGMGYAPAEDDEAEQMHSTG
:***** *** *** ** :. **:***:******* *** * :***:
Mflv_3923|M.gilvum_PYR-GCK VIDPETGLATSVPGPGWTAAFSETLIELAAKRRDIVAITAAMPGPTGLSA
Mvan_2477|M.vanbaalenii_PYR-1 VIDPETGLATSVPGPGWTSTFSEALIRLAGKRRDIVAITAAMPGPTGLSA
MSMEG_2776|M.smegmatis_MC2_155 VIDPETGLPTKASGTSWTSVFSDELISQAAKRRDVVAITAAMPGPTGLSA
TH_2209|M.thermoresistible__bu VIDVETGKPPKASAPGWTSVFSDALVRIGARRRDVVAITAAMPGPTGLTA
Rv2682c|M.tuberculosis_H37Rv PIDPATGQATKVAGPGWTATFSDALIGYAQKRRDIVAITAAMPGPTGLTA
Mb2701c|M.bovis_AF2122/97 PIDPATGQATKVAGPGWTATFSDALIGYAQKRRDIVAITAAMPGPTGLTA
MMAR_2032|M.marinum_M PIDPATGQATKLAGPGWTTTFADALIEYAGKRRDIVAITAAMPGPTGLTA
MUL_3319|M.ulcerans_Agy99 PIDPATGQATKLAGPGWTTTFADALIEYAGKRRDIVAITAAMPGPTGLTA
MAV_3577|M.avium_104 VIDPITGQATKVAGPGWTATFSDALIGYARKRRDIVAITAAMPGPTGLTP
MLBr_01038|M.leprae_Br4923 VMDPTTGQPTKIAAPDWTAIFSDALIGYAMKRRDIVAITAAMPGPTGLTA
MAB_2990c|M.abscessus_ATCC_199 IIDPKTGRAIKASAPGWTSVFSDALIDVASEHRNIVAITAAMPGPTGLSK
:* ** . . ....**: *:: *: . .:*::*************:
Mflv_3923|M.gilvum_PYR-GCK FRKRFPDRFFDVGIAEQHAMTSAAGLAMGGMHPVVALYSTFLNRAFDQML
Mvan_2477|M.vanbaalenii_PYR-1 FRDRFPDRFFDVGIAEQHAMTSAAGLAMGGMHPVVAIYSTFLNRAFDQML
MSMEG_2776|M.smegmatis_MC2_155 FRDRFPDRFFDVGIAEQHAMTSAAGLAMGGLHPVVAIYSTFLNRAFDQLM
TH_2209|M.thermoresistible__bu FRERFPDRFFDVGIAEQHAMTSAAGLAMGGLHPVVAVYSTFLNRAFDQML
Rv2682c|M.tuberculosis_H37Rv FGQRFPDRLFDVGIAEQHAMTSAAGLAMGGLHPVVAIYSTFLNRAFDQIM
Mb2701c|M.bovis_AF2122/97 FGQRFPDRLFDVGIAEQHAMTSAAGLAMGGLHPVVAIYSTFLNRAFDQIM
MMAR_2032|M.marinum_M FGQQFPDRLFDVGIAEQHAMTSAAGLAMGGMHPVVAIYSTFLNRAFDQIM
MUL_3319|M.ulcerans_Agy99 FGQQFPDRLFDVGIAEQHAMTSAAGLAMGGMHPVVAIYSTFLNRAFDQIM
MAV_3577|M.avium_104 FGQQFPDRLFDVGIAEQHAMTSAAGLAMGGMHPVVAIYSTFLNRAFDQIM
MLBr_01038|M.leprae_Br4923 FGQCFPDRLFDVGIAEQHAMTSAAGLAMGRMHPVVAIYSTFLNRAFDQIM
MAB_2990c|M.abscessus_ATCC_199 FGERFPDRLFDVGIAEQHAVTSAAGLAMGGLHPVVAIYSTFLNRAFDQVM
* . ****:**********:********* :*****:***********::
Mflv_3923|M.gilvum_PYR-GCK MDVALHKLPVTMVLDRSGITGPDGASHNGMWDLSILGIVPGMRVAAPRDG
Mvan_2477|M.vanbaalenii_PYR-1 MDVALHKLPVTVVLDRSGVTGPDGASHNGMWDLSILGIVPGMRVAAPRDG
MSMEG_2776|M.smegmatis_MC2_155 MDVALHKLPVTMVLDRAGITGPDGASHNGMWDLSILGIIPGIRVAAPRDG
TH_2209|M.thermoresistible__bu MDVALHKLPVTIVLDRSGITGPDGASHNGMWDLSILGIVPGMRVAAPRDG
Rv2682c|M.tuberculosis_H37Rv MDVALHKLPVTMVLDRAGITGSDGASHNGMWDLSMLGIVPGIRVAAPRDA
Mb2701c|M.bovis_AF2122/97 MDVALHKLPVTMVLDRAGITGSDGASHNGMWDLSMLGIVPGIRVAAPRDA
MMAR_2032|M.marinum_M MDVALHQLPVTMVLDRAGITGSDGPSHNGMWDLSMLGIVPGIRVAAPRDA
MUL_3319|M.ulcerans_Agy99 MDVALHQLPVTMVLDRAGITGSDGPSHNGMWDLSMLGIIPGIRVAAPRDA
MAV_3577|M.avium_104 MDVALHRLPVTMVLDRAGITGSDGASHNGMWDLSILGVVPGMRVAAPRDA
MLBr_01038|M.leprae_Br4923 MDVALHKLPVTMVIDRAGITGSDGPSHNGMWDLSMLGIVPGMRVAAPRDA
MAB_2990c|M.abscessus_ATCC_199 MDVALHRLPVTFVLDRAGITGSDGASHNGMWDLSILNVVPGMRVAAPRDA
******:****.*:**:*:**.**.*********:*.::**:*******.
Mflv_3923|M.gilvum_PYR-GCK ARLREELAEAVEVKDGPTAVRFPKGDVGEDIPAIERRD----GVDVLAVP
Mvan_2477|M.vanbaalenii_PYR-1 ARLREELGEALDVSDGPTAIRFPKGDVGEDIPAIERRG----DVDVLAVP
MSMEG_2776|M.smegmatis_MC2_155 ARLRAELGEALAVNDGPTAIRFPKGDVGEDIPAVQRRD----GVDVLAVP
TH_2209|M.thermoresistible__bu LRLREQLGEALDVKDGPTAIRFPKGDIGEDIPAVERRD----GVDVLSLP
Rv2682c|M.tuberculosis_H37Rv TRLREELGEALDVDDGPTALRFPKGDVGEDISALERRG----GVDVLAAP
Mb2701c|M.bovis_AF2122/97 TRLREELGEALDVDDGPTALRFPKGDVGEDISALERRG----GVDVLAAP
MMAR_2032|M.marinum_M TRLREELGEALDVDDGPTALRFPKGDVGEDIPALERRD----GMDVLAVP
MUL_3319|M.ulcerans_Agy99 TRLREELGEALDVDDGPTALRFPKGDVGEDIPALERRD----GMDVLAVP
MAV_3577|M.avium_104 ARLREELGEALDVDDGPTALRFPKGDVGEDIPAIERRGSGLSGVDVLALP
MLBr_01038|M.leprae_Br4923 IRLREELGEALDVDDGPTAIRFPKGDVCEDIPALKRRS----GVDVLAVP
MAB_2990c|M.abscessus_ATCC_199 SRLREELTEALAVDDGPTALRFPKGEVGEDIPAIERRD----GVDILARP
*** :* **: *.*****:*****:: ***.*::**. .:*:*: *
Mflv_3923|M.gilvum_PYR-GCK ADGLSDDVLIIAVGAFAAMSVAVAERLRDQGIGVTVVDPRWVLPVPAAIN
Mvan_2477|M.vanbaalenii_PYR-1 ADGMSEDVLLVAVGPFAAMALAVADRLRNQGIGVTVVDPRWVLPVPEEIA
MSMEG_2776|M.smegmatis_MC2_155 ATGLTHDVLLVAVGSFAKMALAVAELLRSQGIGVTVVDPRWVLPVPEALA
TH_2209|M.thermoresistible__bu DDGLSADVLLVTVGPFAAMGLAAAERLRNQGIGVTVIDPRWVLPVPQVIT
Rv2682c|M.tuberculosis_H37Rv ADGLNHDVLLVAIGAFAPMALAVAKRLHNQGIGVTVIDPRWVLPVSDGVR
Mb2701c|M.bovis_AF2122/97 ADGLNHDVLLVAIGAFAPMALAVAKRLHNQGIGVTVIDPRWVLPVSDGVR
MMAR_2032|M.marinum_M ASGLNHDVLLIAVGALASMALAVAKRLHNQGIGVTVIDPRWVLPVADGIG
MUL_3319|M.ulcerans_Agy99 ASGLNHDVLLIAVGALASMALAVAKRLHNQGIGVTVIDPRWVLPVADGIG
MAV_3577|M.avium_104 ASGCNHDVLLIGVGAFAPMALAVARRLADQGIGVTVVDPRWVLPVSDSIL
MLBr_01038|M.leprae_Br4923 ATGLAQDVLLVGVGVFASMALAVAKRLHNQGIGVTVIDPRWVLPVCDGVL
MAB_2990c|M.abscessus_ATCC_199 ASGLHADVLLVAVGSFASTALATADRLGKQGIGVTVVDPRWVLPVPPHVV
* ***:: :* :* .:*.* * .*******:******** :
Mflv_3923|M.gilvum_PYR-GCK TLAAAHKLVVTVEDNGGHGGVGSAVSAALRRAEIDVPCRDAALPQAFFDH
Mvan_2477|M.vanbaalenii_PYR-1 TLATRHKLVVTLEDNGGHGGVGSAVSGALRHKEIDVPCRDAALPQEFFAH
MSMEG_2776|M.smegmatis_MC2_155 DLARDHKLVVTVEDNGLHGGIGSSVSAALRHAEIDVPCRDVGVPQQFQDH
TH_2209|M.thermoresistible__bu ELARDHKLVVTVEDNGGHGGVGSAVSAALRAAEVDVPCRDLALPQQFFDH
Rv2682c|M.tuberculosis_H37Rv ELAVQHKLLVTLEDNGVNGGAGSAVSAALRRAEIDVPCRDVGLPQEFYEH
Mb2701c|M.bovis_AF2122/97 ELAVQHKLLVTLEDNGVNGGAGSAVSAALRRAEIDVPCRDVGLPQEFYEH
MMAR_2032|M.marinum_M DLAVAHKLVVTLEDNGVNGGVGSAVSAALRRAEIDVPCRDVGLPQQFFEH
MUL_3319|M.ulcerans_Agy99 DLAVAHKLVVTLEDNGVNGGVGSAVSAALRRAEIGVHCRDVGLPQQFFEH
MAV_3577|M.avium_104 ELAARHKLVVTCEDNGVNGGVGSAVSAALRRAELDVPCRDVGLPQRFYEH
MLBr_01038|M.leprae_Br4923 ELAHTHKLIVTLEDNGVNGGVGAAVSTALRQVEIDTPCRDVGLPQEFYDH
MAB_2990c|M.abscessus_ATCC_199 ELAGQHRLVVTLEDSGVHGGAGSAVTAAMQDADVDIPSRDIGVPQQFLEH
** *:*:** **.* :** *::*: *:: ::. .** .:** * *
Mflv_3923|M.gilvum_PYR-GCK ASRGEVLAEVGLTERTIARQITGWVAALG-ATAGDR--QVSESVD-----
Mvan_2477|M.vanbaalenii_PYR-1 ASRGEVLESVGLTERNIARQITGWVAALG-ASTGDR--EVSEHVD-----
MSMEG_2776|M.smegmatis_MC2_155 ASRGEVLAEVGLTDRHIARQITGWVAAIGTRSAAED--EVGQTLD-----
TH_2209|M.thermoresistible__bu ASRSEVLAEVGLTEQNVARRITGWVAALSPEMAEEQS-QISERLD-----
Rv2682c|M.tuberculosis_H37Rv ASRSEVLADLGLTDQDVARRITGWVAALGTGVCASD--AIPEHLD-----
Mb2701c|M.bovis_AF2122/97 ASRSEVLADLGLTDQDVARRITGWVAALGTGVCASD--AIPEHLD-----
MMAR_2032|M.marinum_M ASRGELLADLGLTDQDVARRVTGWVAALG-ACVSEA--EIKEHLD-----
MUL_3319|M.ulcerans_Agy99 ASRGELLADLRLTDQDVARRVTGWVAALG-ACVSEA--EIKEHLD-----
MAV_3577|M.avium_104 ASRGELLADLALTDQDIARRITGWVAALG-SGVAEA--EIREHLD-----
MLBr_01038|M.leprae_Br4923 ASRSEVLADLGLTDQDVARRITGWVVAFGHCGSGDD--AGQYGPRSSQTM
MAB_2990c|M.abscessus_ATCC_199 ASRGQVLEELGLTDQHIARKITGWVAAMSREIGDSTVNPAAQQVD-----
***.::* .: **:: :**::****.*:. .