For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VALPNLTRDQAVERAALITVDSYQIILDVTDGNGAPGERTFRSTTTVVFDALPGADTVIDISAHTVRRAS LNDQDLDVSGYDEAAGIPLRGLAQRNVVVVDADCHYSNTGEGLHRFVDPVDGETYLYSQFETADAKRMFA CFDQPDLKATFDVRVTAPAHWKVISNGAPLAAANGVHTFATTPRMSTYLVALIAGPYAAWTDTYIDDHGE IPLGIYCRASLAEYMDAERLFTQTKQGFGFYHKHFGLPYAFGKYDQLFVPEFNAGAMENAGAVTFLEDYV FRSKVTRASYERRAETVLHEMAHMWFGDLVTMTWWDDLWLNESFATFASVLCQSEATEFTEAWTTFATVE KSWAYRQDQLPSTHPIAADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLERFLAGLRDYFRTHAFGNA SFDDLLAALEKASGRDLSNWGEQWLKTTGLNTLRPDFEVDAEGRFTRFAVTQSGAAPGAGETRVHRLAVG IYDDDGSKSSGKLVRVHREELDVSGPITNVPALVGVSRGKLILVNDDDLTYCSLRLDERSLQTALDRIAD IAEPLPRTLVWSAAWEMTREAELRARDFVSLVSGGVHAETEVGVAQRLLLQAQTALGCYAEPGWARERGW PQFADRLLELAREAEPGSDHQLAYINSLCSSVLSPRHVQTLGALLEGEPAACGLAGLAVDTDLRWRIVTA LATAGAIDADGPETPRIDAEVQRDPTAAGKRHAAQARAARPQFVVKDEAFTTVVEDDTLANATGRAMIAG IAAPGQGELLKPFARRYFQAIPGVWARRSSEVAQSVVIGLYPHWDISEQGITAAEEFLSDPEVPPALRRL VLEGQAAVQRSLRARNFDADG
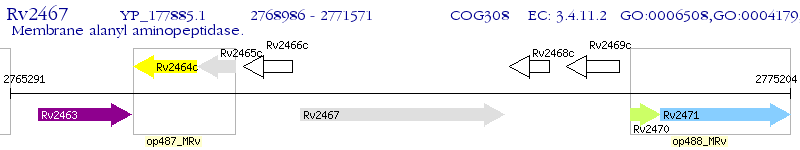
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2467 | pepN | - | 100% (861) | PROBABLE AMINOPEPTIDASE N PEPN (LYSYL AMINOPEPTIDASE) (LYS-AP) (ALANINE AMINOPEPTIDASE) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2494 | pepN | 0.0 | 100.00% (861) | aminopeptidase N |
| M. gilvum PYR-GCK | Mflv_2577 | - | 0.0 | 78.54% (862) | aminopeptidase N |
| M. leprae Br4923 | MLBr_01486 | - | 0.0 | 83.41% (862) | putative aminopeptidase |
| M. abscessus ATCC 19977 | MAB_1571c | - | 0.0 | 78.03% (865) | aminopeptidase |
| M. marinum M | MMAR_3817 | pepN | 0.0 | 85.52% (863) | aminopeptidase N PepN |
| M. avium 104 | MAV_1704 | pepN | 0.0 | 85.40% (863) | aminopeptidase N |
| M. smegmatis MC2 155 | MSMEG_4690 | pepN | 0.0 | 78.19% (862) | aminopeptidase N |
| M. thermoresistible (build 8) | TH_0345 | pepD | 0.0 | 77.20% (864) | PROBABLE AMINOPEPTIDASE N PEPN (LYSYL AMINOPEPTIDASE) |
| M. ulcerans Agy99 | MUL_3742 | pepN | 0.0 | 85.40% (863) | aminopeptidase N PepN |
| M. vanbaalenii PYR-1 | Mvan_4057 | - | 0.0 | 78.56% (863) | aminopeptidase N |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2467|M.tuberculosis_H37Rv ----------------------------MALPNLTRDQAVERAALITVDS
Mb2494|M.bovis_AF2122/97 ----------------------------MALPNLTRDQAVERAALITVDS
MMAR_3817|M.marinum_M ----------------------------MALPNLTRDQAVERAALITVDS
MUL_3742|M.ulcerans_Agy99 ----------------------------MALPNLTRDQAVERAALITVDS
MLBr_01486|M.leprae_Br4923 ----------------------------MALPNLTRDQAVERSALITVDS
MAV_1704|M.avium_104 ----------------------------MALPNLTREQAVERAALVTVAG
MAB_1571c|M.abscessus_ATCC_199 ----------------------------MALPNLTRDQAAARAAAIDVEH
Mflv_2577|M.gilvum_PYR-GCK MSTSGTDDASEQRSAPVLSSRIRLVLASVALPNLTRDQAAERAALVTVDS
Mvan_4057|M.vanbaalenii_PYR-1 ---------------------MRLVLTSVALPNLTRDQAAERAALVTVDS
TH_0345|M.thermoresistible__bu ----------------------------VALPNLTRDQAVERAALVTVDR
MSMEG_4690|M.smegmatis_MC2_155 ----------------------------MALPNLTRDQAIERAALVTVDT
:*******:** *:* : *
Rv2467|M.tuberculosis_H37Rv YQIILDVTDG-NGAPGERTFRSTTTVVFDALPGADTVIDISAHTVRRASL
Mb2494|M.bovis_AF2122/97 YQIILDVTDG-NGAPGERTFRSTTTVVFDALPGADTVIDISAHTVRRASL
MMAR_3817|M.marinum_M YQVNLDVT-------GERTFRSTTTVMFDALPGADTVIDIAAEGVRSASL
MUL_3742|M.ulcerans_Agy99 YQVNLDVT-------GERTFRSTTTVMFDALPGADTVIDIAAEGVRSASL
MLBr_01486|M.leprae_Br4923 YQINLDLTDGPDNAPSDRTFRSITTVVFDALAGADTVIDIAADAVRSATL
MAV_1704|M.avium_104 YRIDLDLTDG-NGAPGERTFRSITTVVFDALAGADTVIDIAADTVRSATL
MAB_1571c|M.abscessus_ATCC_199 YAITLDLTND-DGGPSEETFHSSSTVTFTAQPGTSTVIDIAARTVRRVEL
Mflv_2577|M.gilvum_PYR-GCK YHVSLDLTDG-AGKPGERTFRSVTTVKFDALAGSDTYIDLAADTVRSATL
Mvan_4057|M.vanbaalenii_PYR-1 YRIHLDLTDG-AGKPGERTFRSVTTVEFDAVAGADTYIDLAADSVRSATL
TH_0345|M.thermoresistible__bu YHIELDLTDG-RGNPGAETFRSITTVEFESLPGAETFIDLAARRIRSATL
MSMEG_4690|M.smegmatis_MC2_155 YRIVLDLTDG-QGKPSERTFRSTTTVEFDALAGGETVLDLAADKIHSATL
* : **:* . .**:* :** * : .* .* :*::* :: . *
Rv2467|M.tuberculosis_H37Rv NDQDLDVSGYDEAAGIPLRGLAQRNVVVVDADCHYSNTGEGLHRFVDPVD
Mb2494|M.bovis_AF2122/97 NDQDLDVSGYDEAAGIPLRGLAQRNVVVVDADCHYSNTGEGLHRFVDPVD
MMAR_3817|M.marinum_M NGQELDVSGYDESTGIPLRGLSHRNVVIVDADCRYSNTGEGLHRFVDPVD
MUL_3742|M.ulcerans_Agy99 NGQELDVSGYDESTGIPLRGLSHRNVVIVDADCRYSNTGEGLHRFVDPVD
MLBr_01486|M.leprae_Br4923 NDHELDVSEYDESTGIPLPGLADRNVVVVDADCRYSNTGEGLHRFVDPVD
MAV_1704|M.avium_104 NGRDIDVSGYDESTGIPLQGLADRNVVVVDADCRYSNTGEGLHRFVDPVD
MAB_1571c|M.abscessus_ATCC_199 NGTELDVSGYDEEQGITLPGLAATNTVVVEADCEYSHTGEGLHRFVDPVD
Mflv_2577|M.gilvum_PYR-GCK NGVDIDVSGYDESTGIPLRGLQGHNTLVVDADCRYSNTGEGLHRFVDPVD
Mvan_4057|M.vanbaalenii_PYR-1 NGTEIDVSGYDESTGIPLTGVAGHNVLVVDADCRYSNTGEGLHRFVDPVD
TH_0345|M.thermoresistible__bu NGRPIDVSGYDESTGVPLTVLDKHNVLVVDADCHYSNTGEGLHRFVDPVD
MSMEG_4690|M.smegmatis_MC2_155 NGKPIDVSAYDESTGIPLVGLAEHNVVVVDADCLYSNTGEGLHRFVDPVD
*. :*** *** *:.* : *.::*:*** **:*************
Rv2467|M.tuberculosis_H37Rv GETYLYSQFETADAKRMFACFDQPDLKATFDVRVTAPAHWKVISNGAPLA
Mb2494|M.bovis_AF2122/97 GETYLYSQFETADAKRMFACFDQPDLKATFDVRVTAPAHWKVISNGAPLA
MMAR_3817|M.marinum_M GEAYLYSQFETADAKRMFACFDQPDLKATFEIRVTAPQHWKVVSNAAAVE
MUL_3742|M.ulcerans_Agy99 GEAYLYSQFETADAKRMFACFDQPDLKATFEIRVTAPQHWKVISNAAAVE
MLBr_01486|M.leprae_Br4923 DEIYLYSQFETADAKRMFACFDQPDLKATFEVEVTACKNWKVISNSVCIN
MAV_1704|M.avium_104 DEVYLYSQFETADAKRMFACFDQPDLKATFDVRVTAPRHWKVISNSAAVS
MAB_1571c|M.abscessus_ATCC_199 DEVYLYSQFETADAKRMFACFDQPDLKAAFDVTVTAPAHWQVISNGAALS
Mflv_2577|M.gilvum_PYR-GCK DEVYLYSQFETADAKRMFACFDQPDLKAAFDVTVVAPAHWEVISNGATVD
Mvan_4057|M.vanbaalenii_PYR-1 NEVYLYSQFETADAKRMFACFDQPDLKAFFDVTVVAPGHWEVVSNGATVE
TH_0345|M.thermoresistible__bu GEVYLYSQFETADAKRMFACFDQPDLKAAFDVVVTAPAHWEVISNGAVES
MSMEG_4690|M.smegmatis_MC2_155 DEVYLYSQFETADAKRMFACFDQPDLKATFDVEVTAPTHWEVVSNGATTS
.* ************************* *:: *.* :*:*:**..
Rv2467|M.tuberculosis_H37Rv AAN----G-VHTFATTPRMSTYLVALIAGPYAAWTDTYIDDHGEIPLGIY
Mb2494|M.bovis_AF2122/97 AAN----G-VHTFATTPRMSTYLVALIAGPYAAWTDTYIDDHGEIPLGIY
MMAR_3817|M.marinum_M VSQSQSAA-VHSFGVTPRMSTYLVALVAGPYAVWNDTYSDEHGDIPLGIY
MUL_3742|M.ulcerans_Agy99 VSQSQSAA-VHSFGVTPRMSTYLVALIAGPYAVWNDTYSDEHGDIPLGIY
MLBr_01486|M.leprae_Br4923 ITEG-----THTFATTPRMSTYLVALIAGPYAAWYASYRDEHGEIPLGIY
MAV_1704|M.avium_104 VNDAAQHR-VHTFATTPRMSTYLVALIAGPYAEWKDSYVDEHGEIPLGIY
MAB_1571c|M.abscessus_ATCC_199 VADG-----VHTFATTPKMSTYLVALIAGPYARWNDEYSDEHGTIDLGIY
Mflv_2577|M.gilvum_PYR-GCK ATD-EGGARRHTFKATPRMSTYLVALIAGPYARWDDVYSDDHGDIPLGLF
Mvan_4057|M.vanbaalenii_PYR-1 VEELDSGAKRHTFKATPRMSTYLAALIAGPYARWDDVYSDDHGDIPLGIF
TH_0345|M.thermoresistible__bu IRG-EGPARVHTFVRTPRISTYLVALIAGPYARWDDVYRDEHGEIPLGLF
MSMEG_4690|M.smegmatis_MC2_155 AEV-TGGATVHTFATTPRMSTYLVALIAGPYAVWRDEYTDEYGTIPLGLF
*:* **::****.**:***** * * *::* * **::
Rv2467|M.tuberculosis_H37Rv CRASLAEYMDAERLFTQTKQGFGFYHKHFGLPYAFGKYDQLFVPEFNAGA
Mb2494|M.bovis_AF2122/97 CRASLAEYMDAERLFTQTKQGFGFYHKHFGLPYAFGKYDQLFVPEFNAGA
MMAR_3817|M.marinum_M CRASLAQYMDAERLFTQTKQGFGFYHKHFGIPYAFGKYDQLFVPEFNAGA
MUL_3742|M.ulcerans_Agy99 CRASLAQYMDAERLFTQTKQGFGFYHKHFGIPYAFGKYDQLFVPEFNAGA
MLBr_01486|M.leprae_Br4923 CRASLAQFMDAERLFTQTKKGFNFYHKHFGLPYAFGKYDQLFVPEFNAGA
MAV_1704|M.avium_104 CRASLAQYMDAERLFTQTKQGFGFYHKNFGMPYAFGKYDQLFVPEFNAGA
MAB_1571c|M.abscessus_ATCC_199 CRASLSEFMDADRLFTETKQGFGFYHKNFGTPYAFGKYDQLFVPEFNAGA
Mflv_2577|M.gilvum_PYR-GCK CRRSLADHMDAERLFTETKQGFGFYHNNFGVPYAFGKYDQLFVPEFNAGA
Mvan_4057|M.vanbaalenii_PYR-1 CRKSLAEYMDAERLFTETKQGFAFYHNNFGVPYAFGKYDQLFVPEFNAGA
TH_0345|M.thermoresistible__bu CRRSLAEHMDAERLFTETKQGFGFYHKHFGVPYVFGKYDQLFVPEFNAGA
MSMEG_4690|M.smegmatis_MC2_155 CRKSLAEHMDAERLFTETKQGFGFYHANFGVPYAFGKYDQLFVPEFNAGA
** **::.***:****:**:** *** :** **.****************
Rv2467|M.tuberculosis_H37Rv MENAGAVTFLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMTWWD
Mb2494|M.bovis_AF2122/97 MENAGAVTFLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMTWWD
MMAR_3817|M.marinum_M MENAGAVTFLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMTWWD
MUL_3742|M.ulcerans_Agy99 MENAGAVTFLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMTWWD
MLBr_01486|M.leprae_Br4923 MENAGAVTFLEDYVFRSKVTRASYERRTETVLHEMAHMWFGNLVTMTWWD
MAV_1704|M.avium_104 MENAGAVTLLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMAWWD
MAB_1571c|M.abscessus_ATCC_199 MENAGAVTFLEDYVFRSKVTKYSYERRAETVLHEMAHMWFGDLVTMRWWD
Mflv_2577|M.gilvum_PYR-GCK MENAGAVTFLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMQWWD
Mvan_4057|M.vanbaalenii_PYR-1 MENAGAVTFLEDYVFRSKVTRYSYERRAETVLHEMAHMWFGDLVTMQWWD
TH_0345|M.thermoresistible__bu MENAGAVTFLEDYVFRSKVTRASYERRCETLLHEMAHMWFGDLVTMRWWD
MSMEG_4690|M.smegmatis_MC2_155 MENAGAVTFLEDYVFRSKVTRASYERRAETVLHEMAHMWFGDLVTMRWWD
********:***********: ***** **:**********:**** ***
Rv2467|M.tuberculosis_H37Rv DLWLNESFATFASVLCQSEATEFTEAWTTFATVEKSWAYRQDQLPSTHPI
Mb2494|M.bovis_AF2122/97 DLWLNESFATFASVLCQSEATEFTEAWTTFATVEKSWAYRQDQLPSTHPI
MMAR_3817|M.marinum_M DLWLNESFATFASVLCQSEATEFTEAWTTFATVEKSWAYRQDQLPSTHPI
MUL_3742|M.ulcerans_Agy99 DLWLNESFATFASVLCQSEATEFTEAWTTFATVEKSWAYRQDQLPSTHPI
MLBr_01486|M.leprae_Br4923 DLWLNESFATFASVLCQSEATEFTEAWTTFATVEKSWAYCQDQLPSTHPI
MAV_1704|M.avium_104 DLWLNESFATFASVLCQSEATEFTEAWTTFATVEKSWAYRQDQLPSTHPI
MAB_1571c|M.abscessus_ATCC_199 DLWLNESFATFASVLCQAEATEYTEAWTTFANVEKSWAYRQDQLPSTHPV
Mflv_2577|M.gilvum_PYR-GCK DLWLNESFATFASVLCQAESTEYTQAWTTFANAEKSWAYRQDQLPSTHPV
Mvan_4057|M.vanbaalenii_PYR-1 DLWLNESFATFASVLCQAEATEYTQAWTTFANVEKSWAYRQDQLPSTHPV
TH_0345|M.thermoresistible__bu DLWLNESFATWASVLCQAEATEYTEAWTTFANVEKSWAYRQDQLPSTHPV
MSMEG_4690|M.smegmatis_MC2_155 DLWLNESFATFASVLCQAEATEYKQAWTTFANVEKSWAYRQDQLPSTHPV
**********:******:*:**:.:******..****** *********:
Rv2467|M.tuberculosis_H37Rv AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLERFLAGLRDYFRTHA
Mb2494|M.bovis_AF2122/97 AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLERFLAGLRDYFRTHA
MMAR_3817|M.marinum_M AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLEHFLAGLRDYFRAHA
MUL_3742|M.ulcerans_Agy99 AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLEHFLAGLRDYFRAHA
MLBr_01486|M.leprae_Br4923 VADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLEQFLAGLRDYFRKHA
MAV_1704|M.avium_104 AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLEHFLAGLRDYFRAHA
MAB_1571c|M.abscessus_ATCC_199 AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLENFLAGLRSYFRDHA
Mflv_2577|M.gilvum_PYR-GCK AADIPDLHAVEVNFDGITYAKGASVLKQLVAYVGLDAFLAGLRDYFRDHA
Mvan_4057|M.vanbaalenii_PYR-1 AADIPDLHAVEVNFDGITYAKGASVLKQLVAYVGLDAFLAGLRDYFRDHA
TH_0345|M.thermoresistible__bu AADIPDLAAVEVNFDGITYAKGASVLKQLVAYVGLDEFLAGLREYFRDHA
MSMEG_4690|M.smegmatis_MC2_155 AADIPDLHAVEVNFDGITYAKGASVLKQLVAYVGKEEFLSGLRDYFRDHA
.****** ************************** : **:***.*** **
Rv2467|M.tuberculosis_H37Rv FGNASFDDLLAALEKASGRDLSNWGEQWLKTTGLNTLRPDFEVDAEGRFT
Mb2494|M.bovis_AF2122/97 FGNASFDDLLAALEKASGRDLSNWGEQWLKTTGLNTLRPDFEVDAEGRFT
MMAR_3817|M.marinum_M FGNATFSDLITALEKASGRDLSNWGQQWLKTTGLNTLRPDFDVDADGRFT
MUL_3742|M.ulcerans_Agy99 FGNATFSDLITALEKASGRDLSNWGQQWLKTTGLNTLRPDFDVDADGRFT
MLBr_01486|M.leprae_Br4923 FANATFDDLLVALENASGRDLSHWSQQWLKTTGINMLRPDFDVDADGKFT
MAV_1704|M.avium_104 FGNATFDDLVAALEKASGRDLSDWGRQWLKTTGLNTLRPDFDVDDRGRFT
MAB_1571c|M.abscessus_ATCC_199 FGNATFDDLLGALEKASGRDLSDWGSQWLKTTGLNTLSPDFEVDDEGKFT
Mflv_2577|M.gilvum_PYR-GCK FGNATFGDLLGALEKSSGRDLSGWGRQWLKTTGLNTLRPDFDVDADGTFT
Mvan_4057|M.vanbaalenii_PYR-1 YGNATFGDLLGALEKSSGRDLSGWGRQWLKTTGLNTLRPDFDVDADGRFT
TH_0345|M.thermoresistible__bu FGNATFADLLGALEKASGRDLSGWGQLWLKTTGLNTLRPEFDLGADGRFT
MSMEG_4690|M.smegmatis_MC2_155 FANATFGDLLGALEKASGRDLSGWGQQWLKTTGLNKLRADFDVDSSGAFT
:.**:* **: ***::****** *. ******:* * .:*::. * **
Rv2467|M.tuberculosis_H37Rv RFAVTQSGAAPGAGETRVHRLAVGIYDDD------GSKSSGKLVRVHREE
Mb2494|M.bovis_AF2122/97 RFAVTQSGAAPGAGETRVHRLAVGIYDDD------GSKSSGKLVRVHREE
MMAR_3817|M.marinum_M RFVVQQSGAAPGAGETRVHRLAVGVYDDD------VAGGSGKLVRVHREE
MUL_3742|M.ulcerans_Agy99 RFVVQQSGAALGAGETRVHRLAVGVYDDD------VAGGSGKLVRVHREE
MLBr_01486|M.leprae_Br4923 RFSVMQSGAEPGAGETRVHRLAVGIYDN--------CGEADKLVRVHREE
MAV_1704|M.avium_104 RFAVTQSGAAPGAGETRVHRLVIGIYDDD---------GSGKLVRVHRAE
MAB_1571c|M.abscessus_ATCC_199 RFAVKQSGAAPGAGETRVHRLAVGIYDDAGAADSATSGASGKLVRIHREE
Mflv_2577|M.gilvum_PYR-GCK RFAIEQGGAKPGEGETRVHRLAVGVYDDD---------ASGKLVRVHREE
Mvan_4057|M.vanbaalenii_PYR-1 RFAIEQEGAKPGDGETRVHRLAVGVYDDD---------SSGKLVRVHREE
TH_0345|M.thermoresistible__bu RFTICQSGAQPGAGETRVHRLAVGIYDDD---------GTGKLARIHREE
MSMEG_4690|M.smegmatis_MC2_155 RFTVAQSGAAPGAGETRVHRLAVGIYDD----------VDGKLQRVRREE
** : * ** * ********.:*:**: .** *::* *
Rv2467|M.tuberculosis_H37Rv LDVSGPITNVPALVGVSRGKLILVNDDDLTYCSLRLDERSLQTALDRIAD
Mb2494|M.bovis_AF2122/97 LDVSGPITNVPALVGVSRGKLILVNDDDLTYCSLRLDERSLQTALDRIAD
MMAR_3817|M.marinum_M LDVAGTESEVPALVGVPRGKLILVNDDDLTYCSLRLDGDSLQTALGRIAD
MUL_3742|M.ulcerans_Agy99 LDVAGTESEVPALVGVPRGKLILVNDDDLTYCSLRLDGDSLQTALGRIAD
MLBr_01486|M.leprae_Br4923 LDVEGTETEVPALVGISQGKLILVNDDDLTYCSLRLDAQSLQTALQSIAD
MAV_1704|M.avium_104 LDVEGPVTEVPALVGVSRGKLVLVNDDDLTYCSVRLDAESLGTALDRIAD
MAB_1571c|M.abscessus_ATCC_199 LDVEGPLTDVPALVGVSRGKLVLVNDDDLTYCSLRLDDESLETLLTRIAD
Mflv_2577|M.gilvum_PYR-GCK LDVEGSTTDVPGLHGVSRGKLILVNDDDLTYCSLRLDPDSLQTVLSRIAD
Mvan_4057|M.vanbaalenii_PYR-1 LDVDGSVTEVPALQGVSRGKFILVNDDDLTYCSLRLDPESLQTVLTRIAD
TH_0345|M.thermoresistible__bu LDVEGEYTDVPALQGVSRGKLMLLNDDDLTYCSLRLDPESLQTVLTRIAD
MSMEG_4690|M.smegmatis_MC2_155 LDVEGPRTEVPELVGVSRGQLILVNDDDLTYCSLRLDPDSLSTVVTRIAD
*** * ::** * *:.:*:::*:*********:*** ** * : ***
Rv2467|M.tuberculosis_H37Rv IAEPLPRTLVWSAAWEMTREAELRARDFVSLVSGGVHAETEVGVAQRLLL
Mb2494|M.bovis_AF2122/97 IAEPLPRTLVWSAAWEMTREAELRARDFVSLVSGGVHAETEVGVAQRLLL
MMAR_3817|M.marinum_M IAEPLPRSLVWSAAWEMTREAELRARDFVSLVSGGIHAETEVGVAQRLLL
MUL_3742|M.ulcerans_Agy99 IAEPLPRSLVWSAAWEMTREAELRARDFVSLVSGGIHAETEVGVAQRLLL
MLBr_01486|M.leprae_Br4923 IAEPLPRTLVWSTAWEMTREAELRARDFVALVSGALHAETEVGVAQRLLL
MAV_1704|M.avium_104 IAEPLPRSLVWSAAWEMTREAELRARDFVALVAGGVQGETEVGVAQRLLL
MAB_1571c|M.abscessus_ATCC_199 IAEPLPRTLAWSAAWEMTREAELRARDFVALVSSGVHAESEVGVAQRLLL
Mflv_2577|M.gilvum_PYR-GCK IAEPLPRTLAWSAAWEMTRDAELRARDFVALVISGLHAETEVGVAQRLVM
Mvan_4057|M.vanbaalenii_PYR-1 IADPLPRTLAWSAAWEMTRDAELRARDFVALVIGGLHAESEVGVAQRLLL
TH_0345|M.thermoresistible__bu IAEPLPRTLSWSAAWEMTRDAELKARDFVALVCRGVHAESEVGVLQRLLL
MSMEG_4690|M.smegmatis_MC2_155 IADPLPRTLAWSAAWEMTRDAEMKARDFVALVSSGVHAETEVGVLQRLLM
**:****:* **:******:**::*****:** .::.*:**** ***::
Rv2467|M.tuberculosis_H37Rv QAQTALGCYAEPGWARERGWPQFADRLLELAREAEPGSDHQLAYINSLCS
Mb2494|M.bovis_AF2122/97 QAQTALGCYAEPGWARERGWPQFADRLLELAREAEPGSDHQLAYINSLCS
MMAR_3817|M.marinum_M QAQTALGSYAEPGWASETGWPQFADRLVELARAADAGSDHQLAFINALCT
MUL_3742|M.ulcerans_Agy99 QAQTALGSYAEPGWASETGWPQFADRLVELARAADAESDHQLAFINALCT
MLBr_01486|M.leprae_Br4923 QAQTALGSYADPDWARDYGWPQFADRILELARAAVSGSDHQLAFINTLCS
MAV_1704|M.avium_104 QAQTALSSYAEPEWARDHGWPQFADRLLELARAAETGSDHQLAFVNALCS
MAB_1571c|M.abscessus_ATCC_199 QAQTALSSYAEPAWAREHGWPAFADRLLELARAAEPGSDHQLAFVNALTG
Mflv_2577|M.gilvum_PYR-GCK QAQTALGSYADPAWAAENGWPAFGDALLDLAAQSAPGSDHQLAFVNALCS
Mvan_4057|M.vanbaalenii_PYR-1 QAQTALGSYADPAWAAENGWPAFGDALLDLARESAPGSDHQLAFVNALCT
TH_0345|M.thermoresistible__bu QAQTALTSYAEPGWAAEQGWPEFADRLLGLARAAAPGSDHQLAYVNALCA
MSMEG_4690|M.smegmatis_MC2_155 QAQTALGSYADPEWATTEGWPGFADRLLELARAAEAGSDHQLAYVNALTT
****** .**:* ** *** *.* :: ** : . ******::*:*
Rv2467|M.tuberculosis_H37Rv SVLSPRHVQTLGALLEGEPAACGLAGLAVDTDLRWRIVTALATAGAIDAD
Mb2494|M.bovis_AF2122/97 SVLSPRHVQTLGALLEGEPAACGLAGLAVDTDLRWRIVTALATAGAIDAD
MMAR_3817|M.marinum_M SVLSPRHVETLAALLDNDPAELGLAGLEVDTDLRWRIVTALATAGVIDSD
MUL_3742|M.ulcerans_Agy99 SVLSPRHVETLAALLDNDPAELGLAGLEVDTDLRWRIVTALATAGVIDSD
MLBr_01486|M.leprae_Br4923 SVLSPRHVQTLAALLNDDPTELGLPGVQVDTDLRWRIVTALATAGTLDAN
MAV_1704|M.avium_104 SLLSTRHVVTLADLLDHDPAELGLAGLEIDTDLRWRIVTALAAAGEIDAD
MAB_1571c|M.abscessus_ATCC_199 SVLSAGHTVVLQALLDSDPASLDLPGLTVDTDLRWRIVNALAASGALEPD
Mflv_2577|M.gilvum_PYR-GCK SVLSPNHVAVLSTLLDNEPAAVNLEGLVVDTDLRWRVVTALARAGVIDAD
Mvan_4057|M.vanbaalenii_PYR-1 SVLSPNHVAVLSTLLDNEPAAVNLPGLVLDTDLRWRIVTALARAGVIDAD
TH_0345|M.thermoresistible__bu SVLSARHIEVLEALLDRDPAVEGLDGLVVDAELRWRIVTALAAAGQIDNT
MSMEG_4690|M.smegmatis_MC2_155 SVLSARHTTLLADLLDKDPGSLGLEGLVVDTDLRWRIVIALARSGEIDAD
*:**. * * **: :* .* *: :*::****:* *** :* ::
Rv2467|M.tuberculosis_H37Rv GPETPRIDAEVQRDPTAAGKRHAAQARAARPQFVVKDEAFTTVVEDDTLA
Mb2494|M.bovis_AF2122/97 GPETPRIDAEVQRDPTAAGKRHAAQARAARPQFVVKDEAFTTVVEDDTLA
MMAR_3817|M.marinum_M GPESPRIDAEEKRDPTATGKRSSAHARAARPQFPVKDRAFTTVVEDDTLA
MUL_3742|M.ulcerans_Agy99 GPESPRIDAEEKRDPTATGKRSSAHARAARPQFPVKDRAFTTVVEDDTLA
MLBr_01486|M.leprae_Br4923 GPQTPWIDAEMQRDPTAAGKRHGAQAAAARPQSDIKDKAFTTVVEDDTLT
MAV_1704|M.avium_104 GPATPFIDAEVQRDPTAAGKRHAAQAAAARPQLPVKEQAWTTVVEDDTLA
MAB_1571c|M.abscessus_ATCC_199 --ASVFIDTELERDPTAAGKRQAAQARAARPVAEVKETAWKQVIEDDSLP
Mflv_2577|M.gilvum_PYR-GCK GTATPFIDAEADNDRTAAGRRHAAAAAAARPQAAVKDAAWEQVVEDDTLA
Mvan_4057|M.vanbaalenii_PYR-1 GTQTPFIDAEAANDPTAAGRRQAAAASAARPQAAVKAGAWEQVVEDDTLA
TH_0345|M.thermoresistible__bu GVATPFIDAEAQRDPTAAGQRHARAAAAARPQPAVKEQAWQQVIEDDTLS
MSMEG_4690|M.smegmatis_MC2_155 GPETPFIDAEAQRDPTAAGKRQAAAASAARPQDVVKANAWQQVVEDDTLA
: **:* .* **:*:* . * **** :* *: *:***:*.
Rv2467|M.tuberculosis_H37Rv NATGRAMIAGIAAPGQGELLKPFARRYFQAIPGVWARRSSEVAQSVVIGL
Mb2494|M.bovis_AF2122/97 NATGRAMIAGIAAPGQGELLKPFARRYFQAIPGVWARRSSEVAQSVVIGL
MMAR_3817|M.marinum_M NATGRAIIAGIGAPGQGELLKPFTARYFEVILGVWERRSSEVAQSVVIGL
MUL_3742|M.ulcerans_Agy99 NATGRAIIAGIGAPGQGELLKPFTARYFEVILGVWERRSSEVAQSVVIGL
MLBr_01486|M.leprae_Br4923 NAAGRAIIAGIVAPGQGELLKPFTARYFAAIPGVWTRRSSEVAQTVVIGL
MAV_1704|M.avium_104 NITARSMIAGIAQPGQHELLKPFTPRYFEAIPGVWARRSSEVAQTVVVGL
MAB_1571c|M.abscessus_ATCC_199 NITARSVIAGIVQPGQAELLAPFSGRYFDVIEDVWARRSSEVAQTVVIGL
Mflv_2577|M.gilvum_PYR-GCK NITARAIIGGFAQPGQGEVLAPFRDRYFGAISGVWQRRSSEVAQTVVVGL
Mvan_4057|M.vanbaalenii_PYR-1 NITARAIIGGFVQPGQGELLAPFRERYFGAISGVWERRSSEVAQTVVIGL
TH_0345|M.thermoresistible__bu NVTTRAIVGGFVRPGQAEVLAPFTERYFAAIRGVWQRRSSEVAQTVVIGL
MSMEG_4690|M.smegmatis_MC2_155 NIMTRAIVGGIVQPGQAELLTPFTTQYFAAISGVWARRSSEVAQTVVIGL
* *:::.*: *** *:* ** :** .* .** ********:**:**
Rv2467|M.tuberculosis_H37Rv YPHWDISEQGITAAEEFLSDPE--VPPALRRLVLEGQAAVQRSLRARNFD
Mb2494|M.bovis_AF2122/97 YPHWDISEQGITAAEEFLSDPE--VPPALRRLVLEGQAAVQRSLRARNFD
MMAR_3817|M.marinum_M YPYWDITEEGVAAADRFLSDPE--LAPALRRLVSEGQAAVKRSLRARRFD
MUL_3742|M.ulcerans_Agy99 YPYWDITEEGVAAANRFLADPE--LAPALRRLVSEGQAAVKRSLRARRFD
MLBr_01486|M.leprae_Br4923 YPHWDISDVGIAAAEEFLTAPESKVPPALRRLVLEGQAAVKRALRARMFD
MAV_1704|M.avium_104 YPAWDISDEGIAAADRFLSDPQ--VPAALRRLVLEGQAGVKRALRARRFD
MAB_1571c|M.abscessus_ATCC_199 YPSWDISPEALGLADAFLAKEV---PSALRRLVSEGRAGIVRSLRAREFD
Mflv_2577|M.gilvum_PYR-GCK YPSWEISQDALDAADRFLSDPE--VPPALRRLVLEGRAGVERSLRAREFD
Mvan_4057|M.vanbaalenii_PYR-1 YPSWDISGEALDAADRFLSDPD--VPPALRRLVLEGRAGVERSLKARQFD
TH_0345|M.thermoresistible__bu YPSWDISQAGLDTADRFLADPD--LPPPLRRLVLEGRAGVERALRAREFD
MSMEG_4690|M.smegmatis_MC2_155 YPSWDISQNGLDAADAFLSDPE--VPPALRRLVLEGRAGVERALRARAFD
** *:*: .: *: **: ...***** **:*.: *:*:** **
Rv2467|M.tuberculosis_H37Rv ADG-
Mb2494|M.bovis_AF2122/97 ADG-
MMAR_3817|M.marinum_M AAL-
MUL_3742|M.ulcerans_Agy99 AAL-
MLBr_01486|M.leprae_Br4923 SAD-
MAV_1704|M.avium_104 ATAD
MAB_1571c|M.abscessus_ATCC_199 AGS-
Mflv_2577|M.gilvum_PYR-GCK VT--
Mvan_4057|M.vanbaalenii_PYR-1 VT--
TH_0345|M.thermoresistible__bu AR--
MSMEG_4690|M.smegmatis_MC2_155 VS--