For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NSQNSQIQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFLGVQVDDASANDIMAQLLVLESLDPDRD ITMYINSPGGGFTSLMAIYDTMQYVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLSG VIQGQFSDLEIQAAEIERMRTLMETTLARHTGKDAGVIRKDTDRDKILTAEEAKDYGIIDTVLEYRKLSA QTA*
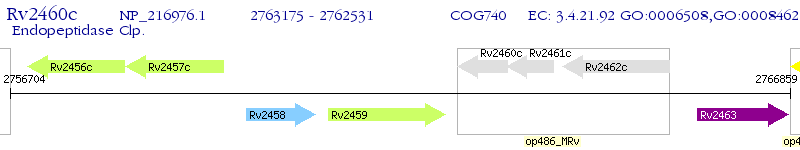
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2460c | clpP2 | - | 100% (214) | ATP-dependent Clp protease proteolytic subunit |
| M. tuberculosis H37Rv | Rv2461c | clpP | 8e-38 | 47.09% (172) | ATP-dependent Clp protease proteolytic subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2487c | clpP2 | 1e-117 | 100.00% (214) | ATP-dependent Clp protease proteolytic subunit |
| M. gilvum PYR-GCK | Mflv_2589 | - | 1e-106 | 91.51% (212) | ATP-dependent Clp protease proteolytic subunit |
| M. leprae Br4923 | MLBr_01479 | clpP2 | 1e-114 | 97.18% (213) | ATP-dependent Clp protease proteolytic subunit |
| M. abscessus ATCC 19977 | MAB_1582 | - | 1e-104 | 90.48% (210) | ATP-dependent Clp protease proteolytic subunit |
| M. marinum M | MMAR_3807 | clpP2 | 1e-112 | 97.14% (210) | ATP-dependent Clp protease proteolytic subunit ClpP2 |
| M. avium 104 | MAV_1714 | clpP | 1e-113 | 97.62% (210) | ATP-dependent Clp protease proteolytic subunit |
| M. smegmatis MC2 155 | MSMEG_4672 | clpP | 1e-109 | 93.40% (212) | ATP-dependent Clp protease proteolytic subunit |
| M. thermoresistible (build 8) | TH_3079 | - | 1e-106 | 92.31% (208) | PUTATIVE Clp protease |
| M. ulcerans Agy99 | MUL_3730 | clpP2 | 1e-112 | 97.14% (210) | ATP-dependent Clp protease proteolytic subunit |
| M. vanbaalenii PYR-1 | Mvan_4039 | - | 1e-108 | 94.23% (208) | ATP-dependent Clp protease proteolytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2460c|M.tuberculosis_H37Rv ------MNSQNSQIQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
Mb2487c|M.bovis_AF2122/97 ------MNSQNSQIQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
MLBr_01479|M.leprae_Br4923 ------MKLQNSQLQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
MAV_1714|M.avium_104 ---------MNPQIQPQARYILPSFIEHSSFGIKESNPYNKLFEERIIFL
MMAR_3807|M.marinum_M ---------MNPQVQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
MUL_3730|M.ulcerans_Agy99 ---------MNPQVQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
Mflv_2589|M.gilvum_PYR-GCK -----MIDHTDPRLQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
Mvan_4039|M.vanbaalenii_PYR-1 --------MTDPRLQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
TH_3079|M.thermoresistible__bu MHNLHLPPHLDPRVQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
MSMEG_4672|M.smegmatis_MC2_155 MSN--IHPSLDARLQPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
MAB_1582|M.abscessus_ATCC_1997 ----MISKHLNPELAPQARYILPSFIEHSSFGVKESNPYNKLFEERIIFL
:..: *****************:*****************
Rv2460c|M.tuberculosis_H37Rv GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGGFTSLMAIYDTMQ
Mb2487c|M.bovis_AF2122/97 GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGGFTSLMAIYDTMQ
MLBr_01479|M.leprae_Br4923 GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGGFTSLMAIYDTMQ
MAV_1714|M.avium_104 GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGGFTSLMAIYDTMQ
MMAR_3807|M.marinum_M GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGGFTSLMAIYDTMQ
MUL_3730|M.ulcerans_Agy99 GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGGFTSLMAIYDTMQ
Mflv_2589|M.gilvum_PYR-GCK GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGSFTSLMAIYDTMQ
Mvan_4039|M.vanbaalenii_PYR-1 GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGSFTSLMAIYDTMQ
TH_3079|M.thermoresistible__bu GVQVDDASANDVMAQLLVLESLDPDRDITMYINSPGGSFTSLMAIYDTMQ
MSMEG_4672|M.smegmatis_MC2_155 GVQVDDASANDIMAQLLVLESLDPDRDITMYINSPGGSFTSLMAIYDTMQ
MAB_1582|M.abscessus_ATCC_1997 GVQVDDASANDIMAQLLVLESLDPDREITMYINSPGGSFTSLMAIYDTMQ
***********:**************:**********.************
Rv2460c|M.tuberculosis_H37Rv YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLSGVIQ
Mb2487c|M.bovis_AF2122/97 YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLSGVIQ
MLBr_01479|M.leprae_Br4923 YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLAGVIQ
MAV_1714|M.avium_104 YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLQGVIQ
MMAR_3807|M.marinum_M YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLSGVIQ
MUL_3730|M.ulcerans_Agy99 YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPSLSGVIQ
Mflv_2589|M.gilvum_PYR-GCK YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPALGGVIQ
Mvan_4039|M.vanbaalenii_PYR-1 YVRADIQTVCLGQAASAAAVLLAAGTPGKRMALPNARILIHQPALGGVIQ
TH_3079|M.thermoresistible__bu YVRADIQTVCLGQAASAAAVLLAAGTKGKRMALPNARVLIHQPALAGVIQ
MSMEG_4672|M.smegmatis_MC2_155 YVRADIQTVCLGQAASAAAVLLAAGTPGKRLALPNARVLIHQPALSGVIQ
MAB_1582|M.abscessus_ATCC_1997 YVRSDIRTVVLGQAASAAAVLLAAGTPGKRMALPNARVLIHQPALSGVIQ
***:**:** **************** ***:******:*****:* ****
Rv2460c|M.tuberculosis_H37Rv GQFSDLEIQAAEIERMRTLMETTLARHTGKDAGVIRKDTDRDKILTAEEA
Mb2487c|M.bovis_AF2122/97 GQFSDLEIQAAEIERMRTLMETTLARHTGKDAGVIRKDTDRDKILTAEEA
MLBr_01479|M.leprae_Br4923 GQFSDLEIQAAEIERMRTLMETTLSRHTGKDAAVIRKDTDRDKILTAEEA
MAV_1714|M.avium_104 GQFSDLEIQAAEIERMRTLMETTLARHTGKDAATIRKDTDRDKILTAEEA
MMAR_3807|M.marinum_M GQFSDLEIQAAEIERMRTLMESTLARHTGKDPSVIRKDTDRDKILTAEEA
MUL_3730|M.ulcerans_Agy99 GQFSDLEIQAAEIERMRTLMESTLARHTGKDPSVIRKDTDRDKILTAEEA
Mflv_2589|M.gilvum_PYR-GCK GQFSDLEIQAAEIERMRTLMEETLSLHTGKPADVIRKDTDRDKILTAAAA
Mvan_4039|M.vanbaalenii_PYR-1 GQFSDLEIQAAEIERMRTLMEETLARHTGKDAAQIRKDTDRDKILTAEQA
TH_3079|M.thermoresistible__bu GQFSDLEIQAAEVERMRTLMEHTLAHHTGKDPETIRRDTDRDKILTAEQA
MSMEG_4672|M.smegmatis_MC2_155 GQFSDLEIQAAEIERMRTLMETTLARHTGKDPAQIRKDTDRDKILTAEEA
MAB_1582|M.abscessus_ATCC_1997 GQFTDLEIQAAEIERMRVLQETTLARHTGRPAEEIRKDTDRDKIFTAEEA
***:********:****.* * **: ***: . **:*******:** *
Rv2460c|M.tuberculosis_H37Rv KDYGIIDTVLEYRKLSAQTA-
Mb2487c|M.bovis_AF2122/97 KDYGIIDTVLEYRKLSAQTA-
MLBr_01479|M.leprae_Br4923 KDYGIIDTVLEYRKLSAQTV-
MAV_1714|M.avium_104 KDYGIIDTVLEYRKLSAQTA-
MMAR_3807|M.marinum_M KDYGIIDTVLEYRKLSAQNA-
MUL_3730|M.ulcerans_Agy99 KDYGIIDTVLEYRKLSAQNA-
Mflv_2589|M.gilvum_PYR-GCK KDYGIIDTVLEYRKLSAQKG-
Mvan_4039|M.vanbaalenii_PYR-1 KEYGIIDTVLEYRKLSAQKA-
TH_3079|M.thermoresistible__bu KEYGIIDTVLEYRKLSAQTA-
MSMEG_4672|M.smegmatis_MC2_155 KEYGIIDTVLQYRKLSAQTS-
MAB_1582|M.abscessus_ATCC_1997 KEYGIIDVVLEYRKLSAQTAS
*:*****.**:*******.