For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRYSAYRRGPDVISPDVIDRILVGACAAVWLVFTGVSVAAAVALMDLGRGFHEMAGNPHTTWVLYAVIV VSALVIVGAIPVLLRARRMAEAEPATRPTGASVRGGRSIGSGHPAKRAVAESAPVQHADAFEVAAEWSSE AVDRIWLRGTVVLTSAIGIALIAVAAATYLMAVGHDGPSWISYGLAGVVTAGMPVIEWLYARQLRRVVAP QSS*
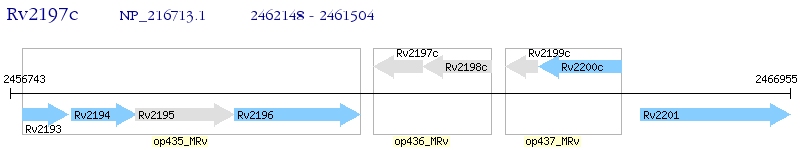
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2197c | - | - | 100% (214) | Probable conserved transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2220c | - | 1e-118 | 99.53% (214) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2952 | - | 5e-35 | 40.76% (211) | hypothetical protein Mflv_2952 |
| M. leprae Br4923 | MLBr_00878 | - | 2e-70 | 62.56% (211) | hypothetical protein MLBr_00878 |
| M. abscessus ATCC 19977 | MAB_1964 | - | 7e-33 | 36.99% (219) | hypothetical protein MAB_1964 |
| M. marinum M | MMAR_3241 | - | 3e-71 | 61.72% (209) | transmembrane protein |
| M. avium 104 | MAV_2294 | - | 1e-73 | 64.95% (214) | hypothetical protein MAV_2294 |
| M. smegmatis MC2 155 | MSMEG_4264 | - | 5e-34 | 41.58% (190) | hypothetical protein MSMEG_4264 |
| M. thermoresistible (build 8) | TH_1050 | glgB | 3e-05 | 55.88% (34) | PROBABLE 1,4-ALPHA-GLUCAN BRANCHING ENZYME GLGB (GLYCOGEN |
| M. ulcerans Agy99 | MUL_3554 | - | 3e-71 | 62.20% (209) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_3561 | - | 7e-38 | 45.15% (206) | hypothetical protein Mvan_3561 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2952|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 --------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2220c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2294|M.avium_104 --------------------------------------------------
MLBr_00878|M.leprae_Br4923 --------------------------------------------------
MMAR_3241|M.marinum_M --------------------------------------------------
MUL_3554|M.ulcerans_Agy99 --------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1050|M.thermoresistible__bu VTSPHLRPDPAELGRLLAGEHHNPHSILGAHEYGDHTVLRAYRPHAQSVV
Mflv_2952|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 --------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2220c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2294|M.avium_104 --------------------------------------------------
MLBr_00878|M.leprae_Br4923 --------------------------------------------------
MMAR_3241|M.marinum_M --------------------------------------------------
MUL_3554|M.ulcerans_Agy99 --------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1050|M.thermoresistible__bu ALVGSDRYPLEHLESGLFAVALPFTGLMDYRLEITYPHSGTHVVADAYRF
Mflv_2952|M.gilvum_PYR-GCK -----------MTHLTDSANRSGAGFDAAALDRTDRLLLGGCAVIWLAAL
Mvan_3561|M.vanbaalenii_PYR-1 -----------MTNLTDTT-RTRRGFDVAALDRTDRILLGAAVVVWLAAL
MSMEG_4264|M.smegmatis_MC2_155 -----------MTSP------------FAELNTTDRILAGACAVAWLAAL
Rv2197c|M.tuberculosis_H37Rv -----------MVSR-YSAYRR--GPDVISPDVIDRILVGACAAVWLVFT
Mb2220c|M.bovis_AF2122/97 -----------MVSR-YSAYRR--GPDVISPDVIDRILVGACAAVWLVFT
MAV_2294|M.avium_104 -----------MVSK-YSAYRRGLGDDTVSPEVIDRILIGACAAIYLALL
MLBr_00878|M.leprae_Br4923 -----------MMNR-YSPYRR--GSDTIASDVIDRILVGVCAAVWLVLI
MMAR_3241|M.marinum_M -----------MLGK-YSAYRRG-PEGVVAPEVVDRVLIGACAAVWLALV
MUL_3554|M.ulcerans_Agy99 -----------MLGK-YSAYRRG-PEGVVAPEVVDRILIGACAAVWLALV
MAB_1964|M.abscessus_ATCC_1997 ---------MPAGRLAKPLEAIAGPLRRSSPESVDRMLIGLCAVIWLAFV
TH_1050|M.thermoresistible__bu LPTLGELDLHLFNEGRHERLWEILGAHPRTFRTADGEVSGVSFAVWAPNA
* : * . . :
Mflv_2952|M.gilvum_PYR-GCK G-------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 G-------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 G-------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv G-------------------------------------------------
Mb2220c|M.bovis_AF2122/97 G-------------------------------------------------
MAV_2294|M.avium_104 G-------------------------------------------------
MLBr_00878|M.leprae_Br4923 G-------------------------------------------------
MMAR_3241|M.marinum_M G-------------------------------------------------
MUL_3554|M.ulcerans_Agy99 G-------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 G-------------------------------------------------
TH_1050|M.thermoresistible__bu KGVSLIGDFNGWSGTDAPMRVLGSSGVWEVFWPDFPVGGLYKFRIHGADG
Mflv_2952|M.gilvum_PYR-GCK --------AAVAAVVALIDLSRGHATSSADSGTPWVLYTVIGVSAAVIVA
Mvan_3561|M.vanbaalenii_PYR-1 --------AGVAATVALVDLSRGHVESAADSGTPWVLYTIIGISAAVIVA
MSMEG_4264|M.smegmatis_MC2_155 --------AGVAALVALVDLGRGHTQAVESTETPWLLYTVIGISVLVIAA
Rv2197c|M.tuberculosis_H37Rv --------VSVAAAVALMDLGRGFHEMAGNPHTTWVLYAVIVVSALVIVG
Mb2220c|M.bovis_AF2122/97 --------VSVAAAVALMDLGRGFHEMAGNPHTTWVLYAVIVVSALVIVG
MAV_2294|M.avium_104 --------VSVAACVALADLGRGFHKAASSPHTTWVLYAVIIVSALIIAG
MLBr_00878|M.leprae_Br4923 --------VSVAAAVALEDLGRGFHKIASDPHTTWVLYGIIVVSVLIIAG
MMAR_3241|M.marinum_M --------VGVAAVVALADLGRGVQKISKDSHSSWVLYAVIVVSALIIVG
MUL_3554|M.ulcerans_Agy99 --------VGVAAVVALADLGRGVKKISKDSHSSWVLYAVIVVSALIIVG
MAB_1964|M.abscessus_ATCC_1997 --------MLVGAIVVIAGMG----EDSGSSGSSWGIYIVIGVSLAVIVG
TH_1050|M.thermoresistible__bu SVTDRADPMAFATEVPPNTASRVTRSEYTWSDDDWMARRALRNPVFEPMS
..: * . . * : . .
Mflv_2952|M.gilvum_PYR-GCK SVPLLLRARRAPDDELT----------------PVEQRLARQAAERT-QT
Mvan_3561|M.vanbaalenii_PYR-1 AVPLLLRARRTAEADAP----------------AAPQRPVTETAP---PP
MSMEG_4264|M.smegmatis_MC2_155 AIPLLLRARAQADSRPP----------------ARSQGPRREPVA----P
Rv2197c|M.tuberculosis_H37Rv AIPVLLRARRMAEAEPA----------------TRPTGASVRGGRS--IG
Mb2220c|M.bovis_AF2122/97 AIPVLLRARRMAEAEPA----------------TRPTGASVRGGRS--IG
MAV_2294|M.avium_104 AIPILLRARRMSQAEPT----------------ARAMTAPARPPVR--LG
MLBr_00878|M.leprae_Br4923 AVPVLLWARRVARVEPP----------------IRPAGVPERGGVRQLVS
MMAR_3241|M.marinum_M AVPMLLRARRMAETGPL----------------VTSVPLRRTGGQP--LR
MUL_3554|M.ulcerans_Agy99 AVPMLLRARRMAETGPL----------------VTSVPLRRTGGQP--LR
MAB_1964|M.abscessus_ATCC_1997 AVPLLLRARKSAAQRTARG--------------ASKTAPKPKGANSSGGA
TH_1050|M.thermoresistible__bu TYEVHLMSWRPGLSYRELATELTEYVVEQGFTHVELLPVAEHPFGGSWGY
: : * :
Mflv_2952|M.gilvum_PYR-GCK PARPVDAPTEKLRVPPPPV-------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 PARGIDAPTEKLRAARP---------------------------------
MSMEG_4264|M.smegmatis_MC2_155 RRVGYTAPTYRQDAVAT---------------------------------
Rv2197c|M.tuberculosis_H37Rv SGHPAKRAVAES-APVQH--------------------------------
Mb2220c|M.bovis_AF2122/97 SGHPAKRAVAES-APVQH--------------------------------
MAV_2294|M.avium_104 AG--IARPATER-APHTP--------------------------------
MLBr_00878|M.leprae_Br4923 AGRSTARIEVERVCAEER--------------------------------
MMAR_3241|M.marinum_M TANATMRAADRS-ARRDH--------------------------------
MUL_3554|M.ulcerans_Agy99 TANATMRAADRS-ARRDH--------------------------------
MAB_1964|M.abscessus_ATCC_1997 TAPSGPPPTEASTEKLKTFGGLADHVSH----------------------
TH_1050|M.thermoresistible__bu QVTSYYAPTSRLGTPDDFRFLVDSLHQAGIGVIIDWVPAHFPKDAWALGR
Mflv_2952|M.gilvum_PYR-GCK -------DTNSGRLPRILQSPVSPAVIDSPAVDRLWLRNTAAVAAAMGTA
Mvan_3561|M.vanbaalenii_PYR-1 -------SEGSGPLPRALRSSAAP---VSAVTDQVFLRCTASIASAMGVA
MSMEG_4264|M.smegmatis_MC2_155 -----------------------------AFVEKVWLRFVLSVTCALGVA
Rv2197c|M.tuberculosis_H37Rv -----------------ADAFEVAAEWSSEAVDRIWLRGTVVLTSAIGIA
Mb2220c|M.bovis_AF2122/97 -----------------ADAFEVAAEWSSEAVDRIWLRGTVVLTSAIGIA
MAV_2294|M.avium_104 -----------------ATPPDVG--WSGEAVDRIWLRGTVILTGTMGAA
MLBr_00878|M.leprae_Br4923 -----------------VQSVAQPGEWFDAAVDRIWLRGTVGLTGTMGAA
MMAR_3241|M.marinum_M -----------------ANGRSEGTQWSSEAVDRVWLRGTVALVGTIGIA
MUL_3554|M.ulcerans_Agy99 -----------------ANGRSEGTEWSSEAVDRVWLRGTVALVGTIGIA
MAB_1964|M.abscessus_ATCC_1997 ------VPKDYNGPGSPAYRAATEAAAKRAVINRLWLRATVGIAGAIGLA
TH_1050|M.thermoresistible__bu FDGTALYEHADPRRGEQLDWGTYVFDFGRPEVRNFLVANALYWLQEFHID
.. : . :
Mflv_2952|M.gilvum_PYR-GCK TLLVSAATYLMAVDSETA-------------------------SWVLYGV
Mvan_3561|M.vanbaalenii_PYR-1 TLLVSVSTYLMAVDSDTA-------------------------AWVLYGV
MSMEG_4264|M.smegmatis_MC2_155 TAVIGLGTYLLATDHSTA-------------------------AWCAYGL
Rv2197c|M.tuberculosis_H37Rv LIAVAAATYLMAVGHDGP-------------------------SWISYGL
Mb2220c|M.bovis_AF2122/97 LIAVAAATYLMAVGHDGP-------------------------SWISYGL
MAV_2294|M.avium_104 LIAVATATYLMAVGHDGS-------------------------SWVGYGF
MLBr_00878|M.leprae_Br4923 LVAVAASTYLMAVGRDGA-------------------------SWVGYVL
MMAR_3241|M.marinum_M LIAVGAATNLMAVGHDGA-------------------------SWVAYGL
MUL_3554|M.ulcerans_Agy99 LIAVGAATNLMAVGHDGA-------------------------SWVAYGL
MAB_1964|M.abscessus_ATCC_1997 LLAVVTGTYLLAVESDTA-------------------------AVIAFVF
TH_1050|M.thermoresistible__bu GLRVDAVASMLYLDYSRPEGGWTPNVHGGRENLEAVQFLQEMNATVHKVA
: : :: . . :
Mflv_2952|M.gilvum_PYR-GCK AGVVTVGMVAIPWYFLRELRAELGS-------------------------
Mvan_3561|M.vanbaalenii_PYR-1 VGVVTVGMVAIPWYFLRELRTDTVS-------------------------
MSMEG_4264|M.smegmatis_MC2_155 AGLVTLGMIALPIIAIRELDRSAA--------------------------
Rv2197c|M.tuberculosis_H37Rv AGVVTAGMPVIEWLYARQLRRVVAPQSS----------------------
Mb2220c|M.bovis_AF2122/97 AGVVTAGMPVIEWLYSRQLRRVVAPQSS----------------------
MAV_2294|M.avium_104 AGVITAAMPVVEWLHIRQLREAVAEQ------------------------
MLBr_00878|M.leprae_Br4923 AGIVTAVMPVIEWIYVRQLRRVG---------------------------
MMAR_3241|M.marinum_M AGIVTAGMPAIEWFYVRQLRRVSVK-------------------------
MUL_3554|M.ulcerans_Agy99 GGIVTAGMPAIEWFYVRQLRRVSVK-------------------------
MAB_1964|M.abscessus_ATCC_1997 AGVITCAMGAIPWVTLKKLRELGSSGS-----------------------
TH_1050|M.thermoresistible__bu PGIVTIAEESTSWPGVTRPTNLGGLGFSMKWNMGWMNDTLEYMRRDPIYR
*::* .
Mflv_2952|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 --------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2220c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2294|M.avium_104 --------------------------------------------------
MLBr_00878|M.leprae_Br4923 --------------------------------------------------
MMAR_3241|M.marinum_M --------------------------------------------------
MUL_3554|M.ulcerans_Agy99 --------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1050|M.thermoresistible__bu SYHHHEMTFSMLYAFSENYVLPISHDEVVHGKGTLWTRMPGDDHAKAAGL
Mflv_2952|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 --------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2220c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2294|M.avium_104 --------------------------------------------------
MLBr_00878|M.leprae_Br4923 --------------------------------------------------
MMAR_3241|M.marinum_M --------------------------------------------------
MUL_3554|M.ulcerans_Agy99 --------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1050|M.thermoresistible__bu RSLLAYQWAHPGKQLLFMGQEFGQRAEWSEERGVDWFQLDEDGFSSGIAQ
Mflv_2952|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 --------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2220c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2294|M.avium_104 --------------------------------------------------
MLBr_00878|M.leprae_Br4923 --------------------------------------------------
MMAR_3241|M.marinum_M --------------------------------------------------
MUL_3554|M.ulcerans_Agy99 --------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1050|M.thermoresistible__bu LVRDINRIYQSRRALWSRDSSHEGYSWIDANDSANNVLSFLRFGDDGSML
Mflv_2952|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4264|M.smegmatis_MC2_155 --------------------------------------------------
Rv2197c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2220c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2294|M.avium_104 --------------------------------------------------
MLBr_00878|M.leprae_Br4923 --------------------------------------------------
MMAR_3241|M.marinum_M --------------------------------------------------
MUL_3554|M.ulcerans_Agy99 --------------------------------------------------
MAB_1964|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1050|M.thermoresistible__bu ACVFNFSGTXXXPGVRRGQPLPADRGRRLPLAAIDQIWLRAGLAIFSAIG
Mflv_2952|M.gilvum_PYR-GCK ------------------------------
Mvan_3561|M.vanbaalenii_PYR-1 ------------------------------
MSMEG_4264|M.smegmatis_MC2_155 ------------------------------
Rv2197c|M.tuberculosis_H37Rv ------------------------------
Mb2220c|M.bovis_AF2122/97 ------------------------------
MAV_2294|M.avium_104 ------------------------------
MLBr_00878|M.leprae_Br4923 ------------------------------
MMAR_3241|M.marinum_M ------------------------------
MUL_3554|M.ulcerans_Agy99 ------------------------------
MAB_1964|M.abscessus_ATCC_1997 ------------------------------
TH_1050|M.thermoresistible__bu AATLLIGAATYLMAGGTRPAPGGAAMPSPG