For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLDYLRDAAEIYRRSFAVIRAEADLARFPADVARVVVRLIHTCGQVDVAEHVAYTDDVVARAGAALAAGA PVLCDSSMVAAGITTSRLPADNQIVSLVADPRATELAARRQTTRSAAGVELCAERLPGAVLAIGNAPTAL FRLLELVDEGAPPPAAVLGGPVGFVGSAQAKEELIERPRGMSYLVVRGRRGGSAMAAAAVNAIASDRE
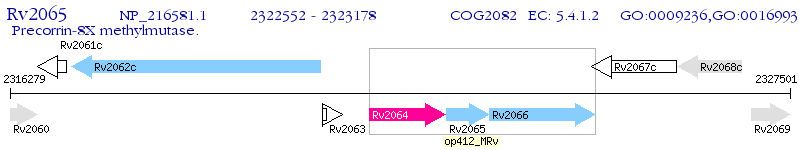
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2065 | cobH | - | 100% (208) | precorrin-8X methylmutase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2091 | cobH | 1e-112 | 100.00% (208) | precorrin-8X methylmutase |
| M. gilvum PYR-GCK | Mflv_3105 | cobH | 9e-92 | 80.29% (208) | precorrin-8X methylmutase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2199c | cobH | 3e-88 | 77.88% (208) | precorrin-8X methylmutase |
| M. marinum M | MMAR_3045 | cobH | 6e-98 | 87.02% (208) | precorrin-8x methylmutase, CobH |
| M. avium 104 | MAV_2431 | - | 6e-82 | 87.28% (173) | precorrin-8X methylmutase |
| M. smegmatis MC2 155 | MSMEG_3872 | cobH | 4e-93 | 83.65% (208) | precorrin-8X methylmutase |
| M. thermoresistible (build 8) | TH_0439 | cobH | 3e-94 | 83.65% (208) | Probable precorrin-8X methylmutase CobH (aka precorrin |
| M. ulcerans Agy99 | MUL_2289 | cobH | 4e-97 | 86.54% (208) | precorrin-8X methylmutase |
| M. vanbaalenii PYR-1 | Mvan_3430 | cobH | 1e-90 | 80.29% (208) | precorrin-8X methylmutase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3105|M.gilvum_PYR-GCK MLDYIRDAAEIYRQSFATIRAEANLARFPEDVARVVVRLIHTCGQVDVAD
Mvan_3430|M.vanbaalenii_PYR-1 MLDYIRDASEIYRQSFATIRAEADLSRFPEDVARVVVRLIHTCGQVDVAE
Rv2065|M.tuberculosis_H37Rv MLDYLRDAAEIYRRSFAVIRAEADLARFPADVARVVVRLIHTCGQVDVAE
Mb2091|M.bovis_AF2122/97 MLDYLRDAAEIYRRSFAVIRAEADLARFPADVARVVVRLIHTCGQVDVAE
MAV_2431|M.avium_104 -----------------------------------MVRLIHTCGQVDVAE
MMAR_3045|M.marinum_M MLDYIRDAAEIYRQSFATIRAEADLSRFPADIARVVVRLIHTCGQVDVAE
MUL_2289|M.ulcerans_Agy99 MLDYIRDAAEIYRQSFATIRAEADLSRFPADIARVVVRLIHTCGQVDVAE
MSMEG_3872|M.smegmatis_MC2_155 MLDYIRDAAEIYRQSFATIRDEADLSRFPADVSRVVVRLIHTCGQVDVTD
TH_0439|M.thermoresistible__bu VLDYIRDAAEIYRQSFATIRAEADLTRFPADVARVVVRLIHTCGQVDLTE
MAB_2199c|M.abscessus_ATCC_199 MLDYVRDGAEIYRQSFATIRAEADLSAFPDDVARVVVRLIHTCGQVDLPG
:***********:.
Mflv_3105|M.gilvum_PYR-GCK HVAFTSDVTARAHAALAAGAPILCDSSMVAAGITRSRLPADNEVVSLVAD
Mvan_3430|M.vanbaalenii_PYR-1 HVAFTRDVTARTHAALTAGAPILCDSSMVAAGITRLRLPADNEVVSLVAD
Rv2065|M.tuberculosis_H37Rv HVAYTDDVVARAGAALAAGAPVLCDSSMVAAGITTSRLPADNQIVSLVAD
Mb2091|M.bovis_AF2122/97 HVAYTDDVVARAGAALAAGAPVLCDSSMVAAGITTSRLPADNQIVSLVAD
MAV_2431|M.avium_104 HVAFTDDVVERVGTALRAGAPVLCDSSMVAAGITAARLPADNEVVSLVAD
MMAR_3045|M.marinum_M HVAYTADVVTRAGTALRDGAPVLCDSSMVAAGITVARLPADNQVVSLVAD
MUL_2289|M.ulcerans_Agy99 HVAYTAGVVTRAGTALRDGAPVLCDSSMVAAGITVARLPADNQVVSLVAD
MSMEG_3872|M.smegmatis_MC2_155 HVAFTGDVVTRTHATLADGAPILCDSSMVAAGITRSRLPADNEVVSLVAD
TH_0439|M.thermoresistible__bu HIAFTDDVVIRAHGALAGGAPILCDSSMVAAGITRSRLPADNEVVSLVAD
MAB_2199c|M.abscessus_ATCC_199 EIAFTPGVVARTRAALDAGAPILCDSSMVAAGITRLRLPADNEVVSLVAD
.:*:* .*. *. :* ***:************ ******::******
Mflv_3105|M.gilvum_PYR-GCK PRAPEMAAKLGSTRSAAAVDLWADRIPGAVFAIGNAPTALFRLLELVDDG
Mvan_3430|M.vanbaalenii_PYR-1 PRAPGLAAELGTTRSAAAVDLWSERIPGAVVAIGNAPTALFRLLELVDDG
Rv2065|M.tuberculosis_H37Rv PRATELAARRQTTRSAAGVELCAERLPGAVLAIGNAPTALFRLLELVDEG
Mb2091|M.bovis_AF2122/97 PRATELAARRQTTRSAAGVELCAERLPGAVLAIGNAPTALFRLLELVDEG
MAV_2431|M.avium_104 PRSAELAARRQTTRSAAGVELWADRLPGAVLAIGNAPTALFRLLELVDDG
MMAR_3045|M.marinum_M PRAAGLAARRETTRSAAGVELWADRLAGAVLAIGNAPTALFRLLELIDEG
MUL_2289|M.ulcerans_Agy99 PRAAGLAARRETTRSAAGVELWADRLAGAVLAIGNAPTALFRLLELLDEG
MSMEG_3872|M.smegmatis_MC2_155 SRAPELAARMGTTRSAAAVDLWADRLGGAVLAIGNAPTALFRLLELLDEG
TH_0439|M.thermoresistible__bu ARAADLAARLGTTRSAAAVDLWAERLGGAVLAIGNAPTALFRLLELLDDG
MAB_2199c|M.abscessus_ATCC_199 PRAAELAQRTGTTRSAAAVDLWADRLPGSVAAIGNAPTALFRILELIDEG
.*:. :* . :*****.*:* ::*: *:* ***********:***:*:*
Mflv_3105|M.gilvum_PYR-GCK VAPPAAVLGGPVGFVGSAQSKQELIDNPRGIPYLLVTGRRGGSAMAAAAV
Mvan_3430|M.vanbaalenii_PYR-1 LAPPASVLGGPVGFVGSAQSKQELIDNPRGMSYLVVTGRRGGSAMAAAAV
Rv2065|M.tuberculosis_H37Rv APPPAAVLGGPVGFVGSAQAKEELIERPRGMSYLVVRGRRGGSAMAAAAV
Mb2091|M.bovis_AF2122/97 APPPAAVLGGPVGFVGSAQAKEELIERPRGMSYLVVRGRRGGSAMAAAAV
MAV_2431|M.avium_104 VGPPAGVLGGPVGFVGSAQSKQELVDNPRGMSYLVVRGRRGGSAMAAAAV
MMAR_3045|M.marinum_M APAPAAVLGGPVGFVGSAQSKQELIDRPRGMSYLVVGGRRGGSAMAAAAV
MUL_2289|M.ulcerans_Agy99 APAPAAVLGGPVGFVGSAQSKQELIDRPRGMSYLVVGGRRGGSAMAAAAV
MSMEG_3872|M.smegmatis_MC2_155 APTPAAVLGGPVGFVGSAQSKQELIERPRGMSYLVVTGRRGGSAMAAAAV
TH_0439|M.thermoresistible__bu APVPAAVLGGPVGFVGSAQSKQELIDRPRGMSYLVVTGRRGGSAMAAAAV
MAB_2199c|M.abscessus_ATCC_199 LAPPVSVLGGPVGFVGSAQSKQELIDRPRGMEYLLVRGRRGGSAMAAAAV
*..*************:*:**::.***: **:* *************
Mflv_3105|M.gilvum_PYR-GCK NSIAAEQE
Mvan_3430|M.vanbaalenii_PYR-1 NSIAAEEE
Rv2065|M.tuberculosis_H37Rv NAIASDRE
Mb2091|M.bovis_AF2122/97 NAIASDRE
MAV_2431|M.avium_104 NAIASDRE
MMAR_3045|M.marinum_M NAIANDSE
MUL_2289|M.ulcerans_Agy99 NAIANDSE
MSMEG_3872|M.smegmatis_MC2_155 NAIASERE
TH_0439|M.thermoresistible__bu NAIASEKE
MAB_2199c|M.abscessus_ATCC_199 NAIASDKD
*:** : :