For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSTLHRTPLATAGLALVVALGGCGGGGGDSRETPPYVPKATTVDATTPAPAAEPLTIASPMFADGAPIPV QFSCKGANVAPPLTWSSPAGAAELALVVDDPDAVGGLYVHWIVTGIAPGSGSTADGQTPAGGHSVPNSGG RQGYFGPCPPAGTGTHHYRFTLYHLPVALQLPPGATGVQAAQAIAQAASGQARLVGTFEG*
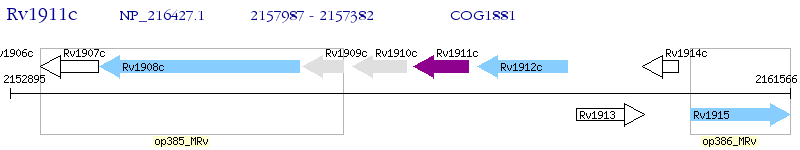
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1911c | lppC | - | 100% (201) | PROBABLE LIPOPROTEIN LPPC |
| M. tuberculosis H37Rv | Rv1910c | - | 8e-67 | 64.32% (199) | hypothetical protein Rv1910c |
| M. tuberculosis H37Rv | Rv2140c | TB18.6 | 2e-12 | 27.75% (173) | hypothetical protein Rv2140c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1946c | lppC | 1e-118 | 100.00% (201) | lipoprotein LppC |
| M. gilvum PYR-GCK | Mflv_3038 | - | 1e-10 | 28.31% (166) | PEBP family protein |
| M. leprae Br4923 | MLBr_01289 | - | 1e-12 | 29.41% (170) | hypothetical protein MLBr_01289 |
| M. abscessus ATCC 19977 | MAB_2103 | - | 4e-11 | 29.59% (169) | hypothetical protein MAB_2103 |
| M. marinum M | MMAR_3122 | - | 4e-12 | 28.32% (173) | hypothetical protein MMAR_3122 |
| M. avium 104 | MAV_2786 | - | 2e-63 | 59.41% (202) | phosphatidylethanolamine-binding protein |
| M. smegmatis MC2 155 | MSMEG_4199 | - | 1e-10 | 26.99% (163) | hypothetical protein MSMEG_4199 |
| M. thermoresistible (build 8) | TH_2369 | - | 3e-12 | 29.45% (163) | CONSERVED HYPOTHETICAL PROTEIN TB18.6 |
| M. ulcerans Agy99 | MUL_2372 | - | 2e-12 | 28.32% (173) | hypothetical protein MUL_2372 |
| M. vanbaalenii PYR-1 | Mvan_3488 | - | 1e-11 | 28.48% (165) | PEBP family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2103|M.abscessus_ATCC_1997 ------------------------------MASP---YDALPQLPTFTLT
MSMEG_4199|M.smegmatis_MC2_155 ------------------------------MTSP---YDNLPQLPSFTLT
MMAR_3122|M.marinum_M ------------------------------MTSP-DPYDALPKLPSFSLT
MUL_2372|M.ulcerans_Agy99 ------------------------------MTSP-DPYDALPKLPSFSLT
MLBr_01289|M.leprae_Br4923 ------------------------------MSLPPDPYAALPKLPSFHLT
TH_2369|M.thermoresistible__bu ------------------------------VAFPYNPYDFLPKLPTFTLT
Mflv_3038|M.gilvum_PYR-GCK ------------------------------MAFDYDPYSFLPELPTFTVT
Mvan_3488|M.vanbaalenii_PYR-1 ------------------------------MAFDYDPYSFLPALPNFTLT
Rv1911c|M.tuberculosis_H37Rv MTSTLHRTP--LATAGLALVVALGGCGGGG-GDSRETPPYVPKATTVDAT
Mb1946c|M.bovis_AF2122/97 MTSTLHRTP--LATAGLALVVALGGCGGGG-GDSRETPPYVPKATTVDAT
MAV_2786|M.avium_104 MEATLVDTSRRIGTIICALALPAGAVGCGGHGHGPTTTPSTPKVTTLGRT
* ... *
MAB_2103|M.abscessus_ATCC_1997 SASVTDGQPLSLDQ--------VSGIMGAGGQDVSPELSWSGFPEETQSF
MSMEG_4199|M.smegmatis_MC2_155 SESVSDGQPLANDQ--------VSGIMGAGGSDISPQLSWSGFPEETKSF
MMAR_3122|M.marinum_M SASITDGQPLATPQ--------VSGIMGAGGEDVSPQLSWSGFPEGTRSF
MUL_2372|M.ulcerans_Agy99 SASITDGQPLATPQ--------VSGIMGAGGEDVSPQLSWSGFPEGTRSF
MLBr_01289|M.leprae_Br4923 SDSITDGQPLATPQ--------ISGIMGAGGADVSPQLSWSGFPTETCSF
TH_2369|M.thermoresistible__bu SESITDGQPLANDQ--------VSGIMGAGGKDISPQLSWSGFPAETRSF
Mflv_3038|M.gilvum_PYR-GCK SQSFEDGGAWGNDQ--------VSGIMGAGGSDISPQLSWSGFPEETRSF
Mvan_3488|M.vanbaalenii_PYR-1 SQSFEDGQPWGKDQ--------VSGIMGAGGSDISPQLSWSGFPEETRSF
Rv1911c|M.tuberculosis_H37Rv TP-APAAEPLTIASPMFADGAPIPVQFSCKGANVAPPLTWS-SPAGAAEL
Mb1946c|M.bovis_AF2122/97 TP-APAAEPLTIASPMFADGAPIPVQFSCKGANVAPPLTWS-SPAGAAEL
MAV_2786|M.avium_104 APNAPAGGPLTITSPAFTDGAPIPPRYTCKGEGIAPPLAWS-APTGA---
: . . . :. . * .::* *:** * :
MAB_2103|M.abscessus_ATCC_1997 AVTVYDPDAPTASGFWHWAVANLPATVTQLPAGVGDG--SGLPGDAITLA
MSMEG_4199|M.smegmatis_MC2_155 AVTIYDPDAPTASGFWHWAVADLPVSVTELPAGAGDG--SPLPGGAVTLA
MMAR_3122|M.marinum_M AVTVYDPDAPTLSGFWHWAVANLPADVTELPAGAGDG--RELPGGALALV
MUL_2372|M.ulcerans_Agy99 AVTVYDPDAPTLSGFWHWAVANLPADVTELPAGAGDG--RELPGGALALV
MLBr_01289|M.leprae_Br4923 AVTVYDPDAPTLSGFWHWAVANLPADVTELPAGVGND--GELPAGALTLI
TH_2369|M.thermoresistible__bu AVTVFDPDAPTASGFWHWAVANLPADVTELPAGAGDG--SPLPGGAITLR
Mflv_3038|M.gilvum_PYR-GCK AVTVYDPDAPTVSGFWHWAVFNLPATVTELPAGVGDGSASGFPGDAVTLA
Mvan_3488|M.vanbaalenii_PYR-1 AVTVFDPDAPTASGFWHWAVFNLPATVTDLPAGVGDGSASGFPGDAVTLA
Rv1911c|M.tuberculosis_H37Rv ALVVDDPDAVGG-LYVHWIVTGIAP-------GSGSTADGQTPAGGHSVP
Mb1946c|M.bovis_AF2122/97 ALVVDDPDAVGG-LYVHWIVTGIAP-------GSGSTADGQTPAGGHSVP
MAV_2786|M.avium_104 ALVVDDPDAPAG-PYVHWVVTGIAP-------GSGSTSAGQTPPGTTTLP
*:.: **** : ** * .:. * *. * . ::
MAB_2103|M.abscessus_ATCC_1997 NDASLKRYIGAAPPAGHGPHRYYIAVHAVSVPKLELPENATPAYLGFNLF
MSMEG_4199|M.smegmatis_MC2_155 NDASVKRYIGAAPPAGHGPHRYYIAVHAVDVEKLDLPENATPAYLGFNLF
MMAR_3122|M.marinum_M NDAGMRRYIGAAPPAGHGVHRYYVAVHAVDVDKLELSEDASPAFLGFNLF
MUL_2372|M.ulcerans_Agy99 NDAGMRRYIGAAPPAGHGVHRYYVAVHAVDVDKLELSEDASPAFLGFNLF
MLBr_01289|M.leprae_Br4923 NDAGMRRYIGAAPPPGHGVHRYYVAVHAVRVQKLNLSEDASPAFLGFNLF
TH_2369|M.thermoresistible__bu NDAGSHRYIGAAPPAGHGFHRYFVAVHALPVEKLDLTEDASPAFLGFNLF
Mflv_3038|M.gilvum_PYR-GCK NDAGLKRFIGAAPPAGHGPHRYIVAVHAVDVEKLDVPADATPAYLGFNLF
Mvan_3488|M.vanbaalenii_PYR-1 NDAGLKRFIGAAPPAGHGVHRYIVAVHAVGVEKLDVPADATPAYLGFNLF
Rv1911c|M.tuberculosis_H37Rv NSGGRQGYFGPCPPAGTGTHHYRFTLYHLPVA-LQLPPGATGVQAAQAIA
Mb1946c|M.bovis_AF2122/97 NSGGRQGYFGPCPPAGTGTHHYRFTLYHLPVA-LQLPPGATGVQAAQAIA
MAV_2786|M.avium_104 NTAGQAGYQGPCPPAGTGTHHYRFTLYQLPND-YQLPAGLAGVQAAQTIG
* .. : *..**.* * *:* .::: : ::. . : . . :
MAB_2103|M.abscessus_ATCC_1997 GHAIARAVIHGTYEQH
MSMEG_4199|M.smegmatis_MC2_155 GHAIARAVIHGTYEQK
MMAR_3122|M.marinum_M QHAIARAVIHGTYEQT
MUL_2372|M.ulcerans_Agy99 QHAIARAVIHGTYEQT
MLBr_01289|M.leprae_Br4923 QHAIARAVIHGTYEQH
TH_2369|M.thermoresistible__bu QHAIARALIHGTYEQK
Mflv_3038|M.gilvum_PYR-GCK GHAIARALITGTYEQN
Mvan_3488|M.vanbaalenii_PYR-1 GHAIARALITGTYEQH
Rv1911c|M.tuberculosis_H37Rv QAASGQARLVGTFEG-
Mb1946c|M.bovis_AF2122/97 QAASGQARLVGTFEG-
MAV_2786|M.avium_104 AAATAQAQLTGVFGG-
* .:* : *.: