For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSPYDNLPQLPSFTLTSESVSDGQPLANDQVSGIMGAGGSDISPQLSWSGFPEETKSFAVTIYDPDAPTA SGFWHWAVADLPVSVTELPAGAGDGSPLPGGAVTLANDASVKRYIGAAPPAGHGPHRYYIAVHAVDVEKL DLPENATPAYLGFNLFGHAIARAVIHGTYEQK*
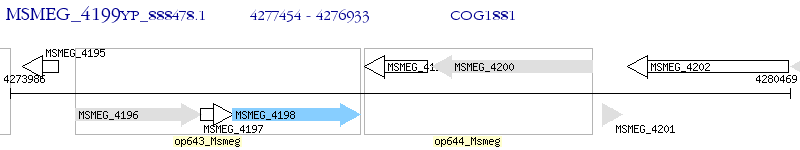
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4199 | - | - | 100% (173) | hypothetical protein MSMEG_4199 |
| M. smegmatis MC2 155 | MSMEG_6851 | - | 4e-06 | 28.31% (166) | phosphatidylethanolamine-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2164c | TB18.6 | 1e-78 | 77.65% (170) | hypothetical protein Mb2164c |
| M. gilvum PYR-GCK | Mflv_3038 | - | 9e-79 | 79.53% (171) | PEBP family protein |
| M. tuberculosis H37Rv | Rv2140c | TB18.6 | 1e-78 | 77.65% (170) | hypothetical protein Rv2140c |
| M. leprae Br4923 | MLBr_01289 | - | 4e-76 | 75.74% (169) | hypothetical protein MLBr_01289 |
| M. abscessus ATCC 19977 | MAB_2103 | - | 5e-88 | 86.05% (172) | hypothetical protein MAB_2103 |
| M. marinum M | MMAR_3122 | - | 1e-80 | 79.31% (174) | hypothetical protein MMAR_3122 |
| M. avium 104 | MAV_2351 | - | 1e-81 | 81.18% (170) | hypothetical protein MAV_2351 |
| M. thermoresistible (build 8) | TH_2369 | - | 4e-84 | 82.46% (171) | CONSERVED HYPOTHETICAL PROTEIN TB18.6 |
| M. ulcerans Agy99 | MUL_2372 | - | 7e-81 | 79.31% (174) | hypothetical protein MUL_2372 |
| M. vanbaalenii PYR-1 | Mvan_3488 | - | 1e-77 | 78.95% (171) | PEBP family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3122|M.marinum_M -MTSPDPYDALPKLPSFSLTSASITDGQPLATPQVSGIMGAGGEDVSPQL
MUL_2372|M.ulcerans_Agy99 -MTSPDPYDALPKLPSFSLTSASITDGQPLATPQVSGIMGAGGEDVSPQL
Mb2164c|M.bovis_AF2122/97 MTTSPDPYAALPKLPSFSLTSTSITDGQPLATPQVSGIMGAGGADASPQL
Rv2140c|M.tuberculosis_H37Rv MTTSPDPYAALPKLPSFSLTSTSITDGQPLATPQVSGIMGAGGADASPQL
MLBr_01289|M.leprae_Br4923 MSLPPDPYAALPKLPSFHLTSDSITDGQPLATPQISGIMGAGGADVSPQL
MAV_2351|M.avium_104 MSTAPDPYASLPKLPTFTLTSQSLTDGGTLAKPQVSGIMGAGGEDVSPQL
TH_2369|M.thermoresistible__bu VAFPYNPYDFLPKLPTFTLTSESITDGQPLANDQVSGIMGAGGKDISPQL
MSMEG_4199|M.smegmatis_MC2_155 MTS---PYDNLPQLPSFTLTSESVSDGQPLANDQVSGIMGAGGSDISPQL
MAB_2103|M.abscessus_ATCC_1997 MAS---PYDALPQLPTFTLTSASVTDGQPLSLDQVSGIMGAGGQDVSPEL
Mflv_3038|M.gilvum_PYR-GCK MAFDYDPYSFLPELPTFTVTSQSFEDGGAWGNDQVSGIMGAGGSDISPQL
Mvan_3488|M.vanbaalenii_PYR-1 MAFDYDPYSFLPALPNFTLTSQSFEDGQPWGKDQVSGIMGAGGSDISPQL
** ** **.* :** *. ** . . *:******** * **:*
MMAR_3122|M.marinum_M SWSGFPEGTRSFAVTVYDPDAPTLSGFWHWAVANLPADVTELPAGAGDG-
MUL_2372|M.ulcerans_Agy99 SWSGFPEGTRSFAVTVYDPDAPTLSGFWHWAVANLPADVTELPAGAGDG-
Mb2164c|M.bovis_AF2122/97 RWSGFPSETRSFAVTVYDPDAPTLSGFWHWAVANLPANVTELPEGVGDG-
Rv2140c|M.tuberculosis_H37Rv RWSGFPSETRSFAVTVYDPDAPTLSGFWHWAVANLPANVTELPEGVGDG-
MLBr_01289|M.leprae_Br4923 SWSGFPTETCSFAVTVYDPDAPTLSGFWHWAVANLPADVTELPAGVGND-
MAV_2351|M.avium_104 SWSGFPAETRSFAVTVYDPDAPTASGFWHWAVANLPADVTELPAGAGDG-
TH_2369|M.thermoresistible__bu SWSGFPAETRSFAVTVFDPDAPTASGFWHWAVANLPADVTELPAGAGDG-
MSMEG_4199|M.smegmatis_MC2_155 SWSGFPEETKSFAVTIYDPDAPTASGFWHWAVADLPVSVTELPAGAGDG-
MAB_2103|M.abscessus_ATCC_1997 SWSGFPEETQSFAVTVYDPDAPTASGFWHWAVANLPATVTQLPAGVGDG-
Mflv_3038|M.gilvum_PYR-GCK SWSGFPEETRSFAVTVYDPDAPTVSGFWHWAVFNLPATVTELPAGVGDGS
Mvan_3488|M.vanbaalenii_PYR-1 SWSGFPEETRSFAVTVFDPDAPTASGFWHWAVFNLPATVTDLPAGVGDGS
***** * *****::****** ******** :**. **:** *.*:.
MMAR_3122|M.marinum_M -RELPGGALALVNDAGMRRYIGAAPPAGHGVHRYYVAVHAVDVDKLELSE
MUL_2372|M.ulcerans_Agy99 -RELPGGALALVNDAGMRRYIGAAPPAGHGVHRYYVAVHAVDVDKLELSE
Mb2164c|M.bovis_AF2122/97 -RELPGGALTLVNDAGMRRYVGAAPPPGHGVHRYYVAVHAVKVEKLDLPE
Rv2140c|M.tuberculosis_H37Rv -RELPGGALTLVNDAGMRRYVGAAPPPGHGVHRYYVAVHAVKVEKLDLPE
MLBr_01289|M.leprae_Br4923 -GELPAGALTLINDAGMRRYIGAAPPPGHGVHRYYVAVHAVRVQKLNLSE
MAV_2351|M.avium_104 -SNLPGDALTLVNDAGQRRYIGAAPPPGHGPHRYYIAVHAVDTEKLDLPE
TH_2369|M.thermoresistible__bu -SPLPGGAITLRNDAGSHRYIGAAPPAGHGFHRYFVAVHALPVEKLDLTE
MSMEG_4199|M.smegmatis_MC2_155 -SPLPGGAVTLANDASVKRYIGAAPPAGHGPHRYYIAVHAVDVEKLDLPE
MAB_2103|M.abscessus_ATCC_1997 -SGLPGDAITLANDASLKRYIGAAPPAGHGPHRYYIAVHAVSVPKLELPE
Mflv_3038|M.gilvum_PYR-GCK ASGFPGDAVTLANDAGLKRFIGAAPPAGHGPHRYIVAVHAVDVEKLDVPA
Mvan_3488|M.vanbaalenii_PYR-1 ASGFPGDAVTLANDAGLKRFIGAAPPAGHGVHRYIVAVHAVGVEKLDVPA
:*..*::* ***. :*::*****.*** *** :****: . **::.
MMAR_3122|M.marinum_M DASPAFLGFNLFQHAIARAVIHGTYEQT
MUL_2372|M.ulcerans_Agy99 DASPAFLGFNLFQHAIARAVIHGTYEQT
Mb2164c|M.bovis_AF2122/97 DASPAYLGFNLFQHAIARAVIFGTYEQR
Rv2140c|M.tuberculosis_H37Rv DASPAYLGFNLFQHAIARAVIFGTYEQR
MLBr_01289|M.leprae_Br4923 DASPAFLGFNLFQHAIARAVIHGTYEQH
MAV_2351|M.avium_104 DASPAFLGFNLFQHAIARAVIHGTYEQK
TH_2369|M.thermoresistible__bu DASPAFLGFNLFQHAIARALIHGTYEQK
MSMEG_4199|M.smegmatis_MC2_155 NATPAYLGFNLFGHAIARAVIHGTYEQK
MAB_2103|M.abscessus_ATCC_1997 NATPAYLGFNLFGHAIARAVIHGTYEQH
Mflv_3038|M.gilvum_PYR-GCK DATPAYLGFNLFGHAIARALITGTYEQN
Mvan_3488|M.vanbaalenii_PYR-1 DATPAYLGFNLFGHAIARALITGTYEQH
:*:**:****** ******:* *****