For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVNRRVPRVRDLAPLLQFNRPQFDTSKRRLGAALTIQDLRRIAKRRTPRAAFDYADGGAEDELSIARARQ GFRDIEFHPTILRDVTTVCAGWNVLGQPTVLPFGIAPTGFTRLMHTEGEIAGARAAAAAGIPFSLSTLAT CAIEDLVIAVPQGRKWFQLYMWRDRDRSMALVRRVAAAGFDTMLVTVDVPVAGARLRDVRNGMSIPPALT LRTVLDAMGHPRWWFDLLTTEPLAFASLDRWPGTVGEYLNTVFDPSLTFDDLAWIKSQWPGKLVVKGIQT LDDARAVVDRGVDGIVLSNHGGRQLDRAPVPFHLLPHVARELGKHTEILVDTGIMSGADIVAAIALGARC TLIGRAYLYGLMAGGEAGVNRAIEILQTGVIRTMRLLGVTCLEELSPRHVTQLRRLGPIGAPT*
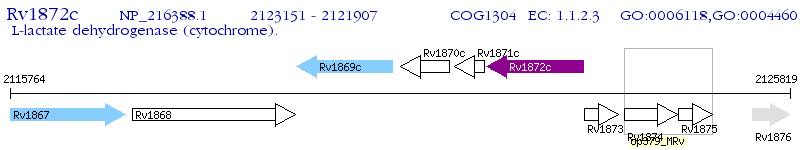
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1872c | lldD2 | - | 100% (414) | POSSIBLE L-LACTATE DEHYDROGENASE (CYTOCHROME) LLDD2 |
| M. tuberculosis H37Rv | Rv0694 | lldD1 | 4e-53 | 35.06% (385) | L-lactate dehydrogenase (cytochrome) LldD1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1903c | lldD2 | 0.0 | 99.52% (414) | L-lactate dehydrogenase (cytochrome) LldD2 |
| M. gilvum PYR-GCK | Mflv_5059 | - | 6e-57 | 33.33% (387) | (S)-2-hydroxy-acid oxidase |
| M. leprae Br4923 | MLBr_02046 | lldD2 | 0.0 | 84.99% (413) | L-lactate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3834c | - | 9e-52 | 33.42% (389) | L-lactate dehydrogenase LldD1 |
| M. marinum M | MMAR_2752 | lldD2 | 0.0 | 87.20% (414) | L-lactate dehydrogenase (cytochrome) LldD2 |
| M. avium 104 | MAV_2843 | - | 0.0 | 85.85% (410) | LldD2 protein |
| M. smegmatis MC2 155 | MSMEG_1424 | - | 3e-53 | 32.91% (392) | FMN-dependent dehydrogenase |
| M. thermoresistible (build 8) | TH_2300 | lldD2 | 1e-174 | 76.96% (395) | POSSIBLE L-LACTATE DEHYDROGENASE (CYTOCHROME) LLDD2 |
| M. ulcerans Agy99 | MUL_3002 | lldD2 | 0.0 | 86.71% (414) | L-lactate dehydrogenase (cytochrome) LldD2 |
| M. vanbaalenii PYR-1 | Mvan_1297 | - | 1e-53 | 32.90% (383) | (S)-2-hydroxy-acid oxidase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1872c|M.tuberculosis_H37Rv MAVNRRVPRVRDLAPLLQFNRPQFDTSKRRLGAALTIQDLRRIAKRRTPR
Mb1903c|M.bovis_AF2122/97 MAVNRRVPRVRDLAPLLQFNRPQFDTSKRRLGAALTIQDLRRIAKRRTPR
MMAR_2752|M.marinum_M MAVDRRLPRVGDLAPLMQFKRPQFNRTKRRLEAALTIEDLRRVAKRRTPR
MUL_3002|M.ulcerans_Agy99 MAVDRRLPRVGDLAPLMQFKRPQFNRTKRRLETALTIEDLRRVAKRRTPR
MAV_2843|M.avium_104 MGVQRRVPRVRDLAPLMQFKRPEFNATKRRLRSAHTIEDLRRIAKRRTPK
MLBr_02046|M.leprae_Br4923 MAAKRRMPRVHDLAPLIQFKRPKFGATKRRLEAALTIQDLRCIAKQRTPK
TH_2300|M.thermoresistible__bu ------------LTGLLGFSAPRL-TTRRRLAAALTIEDLRRLARRRTPR
Mflv_5059|M.gilvum_PYR-GCK -------------------------MARDTWFETVAIAQQR--AKKRLPK
Mvan_1297|M.vanbaalenii_PYR-1 -------------------------MARDTWFETVAIAQQR--AKKRLPK
MSMEG_1424|M.smegmatis_MC2_155 -----------------------MNMARDIWFETVAIAQQR--ARKRLPK
MAB_3834c|M.abscessus_ATCC_199 -------------------------MARNKWFETVAIAQQR--AKKRLPK
:: : :* : * *::* *:
Rv1872c|M.tuberculosis_H37Rv AAFDYADGGAEDELSIARARQGFRDIEFHPTILRDVTTVCAGWNVLGQPT
Mb1903c|M.bovis_AF2122/97 AAFDYADGAAEDELSIARARQGFRDIEFHPTILRDVTTVCAGWNVLGQPT
MMAR_2752|M.marinum_M AAFDYTDGAAEDELSIERARQAFRDIEFHPTILRDVTSVRTGWDVFGKPV
MUL_3002|M.ulcerans_Agy99 AAFDYTDGAAEDELSIERARQAFRDIEFHPTILRDVTSVRTGWDVFGKPV
MAV_2843|M.avium_104 AAFDYTDGAAEDELSIQRARQAFRDIEFHPTILRDVSTVTAGWDVLGGPV
MLBr_02046|M.leprae_Br4923 AAFDYTDGAAEDELSIKRAQQAFRDIEFHPAILRDVTNVCAGWDVLGHSV
TH_2300|M.thermoresistible__bu AAFDYADGAAEEELSLRRARQAFRDIEFHPMILRDVATVRTGWDVLGAPV
Mflv_5059|M.gilvum_PYR-GCK SAYSSLISASEKGVSVSDNVEAFAELGFAPHVVGATDKRDMATTVMGQDI
Mvan_1297|M.vanbaalenii_PYR-1 SAYSSLISASEKGVTVTDNVESFAELGFAPHVIGATEKRDMATSVLGQDI
MSMEG_1424|M.smegmatis_MC2_155 SVYSSLISASEKGVTVTDNVESFAELGFAPHVVGAPEKRDMATTVMGQQI
MAB_3834c|M.abscessus_ATCC_199 SVYSSLISASEKGLTVSDNVEAFGELGFEPHVVGIQPDRELSTTVLGQDI
:.:. ..:*. ::: :.* :: * * :: . *:*
Rv1872c|M.tuberculosis_H37Rv VLPFGIAPTGFTRLMHTEGEIAGARAAAAAGIPFSLSTLATCAIEDLVIA
Mb1903c|M.bovis_AF2122/97 VLPFGIAPTGFTRLMHTEGEIAGARAAAAAGIPFSLSTLATCAIEDLVIA
MMAR_2752|M.marinum_M ALPFGIAPTGFTRLMHTEGEIAGARAAAEAGIPFSMSTLATCAIEDLQAA
MUL_3002|M.ulcerans_Agy99 ALPFGIAPTGFTRLMHTEGEIAGARAAAEAGIPFSMSTLATCAIEDLQAA
MAV_2843|M.avium_104 ALPFGIAPTGFTRLMHTEGEIAGVRAAARAGIPFSLSTLGTCAIEDLAAA
MLBr_02046|M.leprae_Br4923 LLPFGIAPTGFTRLMHTEGEIAGARAAAAAGIPFSLSTLATSAIEDVVAA
TH_2300|M.thermoresistible__bu ALPFGIAPTGFTRLMHTEGERAGARVAGRTGIPFSLSTLATTSIETVKSE
Mflv_5059|M.gilvum_PYR-GCK PLPVIISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKPMEEVTAV
Mvan_1297|M.vanbaalenii_PYR-1 SLPVIISPTG-VQAIDPDGEVAVARAAAARGTAMGLSSFASKPMEDVTAV
MSMEG_1424|M.smegmatis_MC2_155 SLPVIISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKPIEEVVAV
MAB_3834c|M.abscessus_ATCC_199 SLPVMISPTG-VQAVDPDGEVAVARAAAARGTAMGLSSFASKPIEDVVAA
**. *:*** .: :..:** * .*.*. * .:.:*::.: .:* :
Rv1872c|M.tuberculosis_H37Rv VPQGRKWFQLYMWRDRDRSMALVRRVAAAGFDTMLVTVDVPVAGARLRDV
Mb1903c|M.bovis_AF2122/97 VPQGRKWFQLYMWRDRDRSMALVRRAAAAGFDTMLVTVDVPVAGARLRDV
MMAR_2752|M.marinum_M VPQGRKWFQLYMWRDRDRSMALVKRAADAGFDTMLVTVDVPVAGARLRDV
MUL_3002|M.ulcerans_Agy99 VPQGRKWFQLYMWRDRDRSMALVKRAADAGFDTMLVTVDVPVAGARLRDV
MAV_2843|M.avium_104 VPQSRKWFQLYMWKDRERSMALVRRAADAGFDTLLATVDVPVSGARLRDN
MLBr_02046|M.leprae_Br4923 VPQGRKWFQLYMWRDRDRSMALVERAADAGYDALLVTVDVPVAGARLRDT
TH_2300|M.thermoresistible__bu NPHGRNWFQLYMWKDRDRSMALVRRALAAGYDTLLVTVDVPVAGARLRDT
Mflv_5059|M.gilvum_PYR-GCK N--DKIFFQIYWLGSRDAIAARMERARAAGAKGLILTTDWSFSHGRDWGS
Mvan_1297|M.vanbaalenii_PYR-1 N--DKIFFQIYWLGSRDDILARMERARAAGAKGLILTTDWSFAHGRDWGS
MSMEG_1424|M.smegmatis_MC2_155 N--DKIFFQIYWLGDRDAILARAERAKAAGAVGLIVTTDWSFSHGRDWGS
MAB_3834c|M.abscessus_ATCC_199 N--PKTHFQIYWLGGRDDVAQRIQRAKDAGAVGLIATLDWSFSHGRDWGS
: **:* .*: .*. ** :: * * ..: .* .
Rv1872c|M.tuberculosis_H37Rv -RNGMSIPPALTLRTVLDAMGHPRWWFDLLT-------TEPLAFASLDRW
Mb1903c|M.bovis_AF2122/97 -RNGMSIPPALTLRTVLDAMGHPRWWFDLLT-------TEPLAFASLDRW
MMAR_2752|M.marinum_M -RNGMSIPPALTLRTVLDALPHPHWWFDLLT-------TEPLAFASLDRW
MUL_3002|M.ulcerans_Agy99 -RNGMSIPPALTLRTVLDALPHPHWWFDLLT-------TEPLAFASLDRW
MAV_2843|M.avium_104 -RNGMTIPPTLTLRTVLDAVPHPKWWFDLLT-------TEPLAFASLDRW
MLBr_02046|M.leprae_Br4923 -RNGMSIPPALTLRTVFDALPHPRWWIDLLT-------TEPLAFASLDRW
TH_2300|M.thermoresistible__bu -RNGMSIPPALTWRTVLDAAARPRWWFDLLT-------TEPLTFAEGDQW
Mflv_5059|M.gilvum_PYR-GCK PKIPEQMDLKTILKMSPEVLTKPRWFFSFAKTLRPPDLRVPNQGRPGEPG
Mvan_1297|M.vanbaalenii_PYR-1 PKIPERMDLKTMIRMSPEVITKPRWFFSFAKHLRPPDLRVPNQGRRGEAG
MSMEG_1424|M.smegmatis_MC2_155 PKIPEKMDLKTMVTMMPEALTKPRWLWQWGKTMRPPNLRVPNQGARGEDG
MAB_3834c|M.abscessus_ATCC_199 PAIPEKMDLRSMIRLAPEVVTKPSWLWSFGKGMNIPDLRVPNQAARGEAG
: :. :* * . . * :
Rv1872c|M.tuberculosis_H37Rv PGTVGEYLNTVFDPSLTFDDLAWIKSQWPGKLVVKGIQTLDDARAVVDRG
Mb1903c|M.bovis_AF2122/97 PGTVGEYLNTVFDPSLTFDDLAWIKSQWPGKLVVKGIQTLDDARAVVDRG
MMAR_2752|M.marinum_M SGTVGEYLNTMFDPSVTFDDLEWMKAQWPGKLVVKGIQTLDDARAVVERG
MUL_3002|M.ulcerans_Agy99 SGTVGEYLNTMFDPSVTFDDLEWMKAQWPGKLVVKGIQTLDDARAVVERG
MAV_2843|M.avium_104 PGTVAEYLSTMFDPSLTFDDLEWIKARWPGKLVVKGIQTLDDARAVVDRG
MLBr_02046|M.leprae_Br4923 SGTVSDYLNTMFDPSVTFDDLAWIKTQWPGKFVVKGIQTLDDARAVVERG
TH_2300|M.thermoresistible__bu PGTVAEYLDSTFDPTVTFEDLDWIRQHWPNKLVVKGIQTLADARAVVGRG
Mflv_5059|M.gilvum_PYR-GCK PTFFEAYGEWMGTPPPTWEDVAWLREQWGGPFLLKGLVRVDDAKRAVDAG
Mvan_1297|M.vanbaalenii_PYR-1 PTFFEAYGQWMGTPPPTWEDVAWLREQWGGPFLLKGTVRVDDAKRAVDAG
MSMEG_1424|M.smegmatis_MC2_155 PPFFQAYGEWMGTPPPTWEDIAWLREQWDGPFMLKGVIRVDDAKRAVDAG
MAB_3834c|M.abscessus_ATCC_199 PPFFDAYGQWMGTPAPTWDDVRWMREQWDGPFMLKGVMRIDDAKRAVDCG
. . * . *. *::*: *:: :* . :::** : **: .* *
Rv1872c|M.tuberculosis_H37Rv VDGIVLSNHGGRQLDRAPVPFHLLPHVARELGKHTEILVDTGIMSGADIV
Mb1903c|M.bovis_AF2122/97 VDGIVLSNHGGRQLDRAPVPFHLLPHVARELGKHTEILVDTGIMSGADIV
MMAR_2752|M.marinum_M ADGIVLSNHGGRQLDRAPVPFHLLPLVARELGKDTEIVVDTGIMSGADIV
MUL_3002|M.ulcerans_Agy99 ADGIVLSNHGGRQLDRAPAPFHLLPLVARELGKDTEIVVDTGIMSGADIV
MAV_2843|M.avium_104 ADGIVLSNHGGRQLDRAPVPFHLLPTVARELGKHTEILLDTGIMSGADIV
MLBr_02046|M.leprae_Br4923 IDGVVLSNHGGRQLDRAPVPFHLLPTVAREFGKDTEILLDTGIMSGADIV
TH_2300|M.thermoresistible__bu VDGIVLSNHGGRQLDRAPVPLDLLPTVARELGRDTEILLDTGIMSGADIV
Mflv_5059|M.gilvum_PYR-GCK VSAITVSNHGGNNLDGTPAAIRCLPAIADAVGDQVEVLLDGGIRRGSDVV
Mvan_1297|M.vanbaalenii_PYR-1 VSAITVSNHGGNNLDGTPAAIRCLPAIADAVGDQVEVLLDGGIRRGSDVV
MSMEG_1424|M.smegmatis_MC2_155 VSAISVSNHGGNNLDGTPAAIRALPVIAEAVGDQVEVLLDGGIRRGSDVV
MAB_3834c|M.abscessus_ATCC_199 VSAISVSNHGGNNLDGTPASIRALPGIADAVGHDIEVLLDGGIRRGSDVV
..: :*****.:** :*..: ** :* .* . *:::* ** *:*:*
Rv1872c|M.tuberculosis_H37Rv AAIALGARCTLIGRAYLYGLMAGGEAGVNRAIEILQTGVIRTMRLLGVTC
Mb1903c|M.bovis_AF2122/97 AAIALGARCTLIGRAYLYGLMAGGEAGVNRAIEILQTGVIRTMRLLGVTC
MMAR_2752|M.marinum_M AAIALGARCTLIGRAYLYGLMAGGEAGVKRAIEILSAGVSRTMRLLGVTC
MUL_3002|M.ulcerans_Agy99 AAIALGARCTLIGRAYLYGLMAGGEAGVKRAIEILSAGVSRTMRLLGVTC
MAV_2843|M.avium_104 AAIALGARCTLVGRAYLYGLMAGGEAGVTRAIEILAEGVIRTMRLLGVTC
MLBr_02046|M.leprae_Br4923 AAIALGARCTLVGRAYLYGLMAGGEAGVRRAIEILESGVIRTMQLLGVTC
TH_2300|M.thermoresistible__bu AAIALGARFTLVGRAYLYGLMAGGEAGVQRAVEILSSQVTRTMKLLGVSC
Mflv_5059|M.gilvum_PYR-GCK KAVALGARAVMIGRAYLWGLAANGQAGVENVLDILSGGIDSALRGLGKSS
Mvan_1297|M.vanbaalenii_PYR-1 KAVALGARAVMIGRAYLWGLAANGQAGVENVLDILRGGIDSALMGLGKSS
MSMEG_1424|M.smegmatis_MC2_155 KAVALGARAVMIGRAYLWGLAAEGQVGVENVLDILRGGIDSALMGLGRSS
MAB_3834c|M.abscessus_ATCC_199 KALALGARAVMIGRAYLWGLAASGQAGVENVLDIMRGGIDSALMGLGKKS
*:***** .::*****:** * *:.** ..::*: : :: ** ..
Rv1872c|M.tuberculosis_H37Rv LEELSPRHVTQLRRLGPIG------APT
Mb1903c|M.bovis_AF2122/97 LEELSPRHVTQLRRLGPIG------APT
MMAR_2752|M.marinum_M LEELSAKHVTQLQRLGPIP------LPT
MUL_3002|M.ulcerans_Agy99 LEELSAKHVTQLQRLGPIP------LPT
MAV_2843|M.avium_104 LEELSPRHVTQLRRLAPLPPVEPPVTPA
MLBr_02046|M.leprae_Br4923 LEELSPRHVTQLQRLGPIEP------PA
TH_2300|M.thermoresistible__bu LEELSARHATRLHRVAGASDNHHSSSQQ
Mflv_5059|M.gilvum_PYR-GCK IQELTPEDILVPEGFTRTLGV-------
Mvan_1297|M.vanbaalenii_PYR-1 IHELTREDILIPDGFTRTLGA-------
MSMEG_1424|M.smegmatis_MC2_155 IHDLVPEDILVPEGFTRALGVPPASGS-
MAB_3834c|M.abscessus_ATCC_199 VHELSPDDLLIPEGFARGLGRH------
:.:* . .