For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSAPDSPALAGMSIGAVLDLLRPDFPDVTISKIRFLEAEGLVTPRRASSGYRRFTAYDCARLRFILTAQR DHYLPLKVIRAQLDAQPDGELPPFGSPYVLPRLVPVAGDSAGGVGSDTASVSLTGIRLSREDLLERSEVA DELLTALLKAGVITTGPGGFFDEHAVVILQCARALAEYGVEPRHLRAFRSAADRQSDLIAQIAGPLVKAG KAGARDRADDLAREVAALAITLHTSLIKSAVRDVLHR
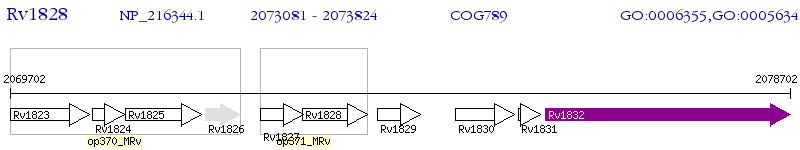
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1828 | - | - | 100% (247) | hypothetical protein Rv1828 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1859 | - | 1e-136 | 100.00% (247) | hypothetical protein Mb1859 |
| M. gilvum PYR-GCK | Mflv_3401 | - | 3e-97 | 75.30% (251) | MerR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02075 | - | 1e-120 | 87.25% (251) | hypothetical protein MLBr_02075 |
| M. abscessus ATCC 19977 | MAB_2404 | - | 2e-91 | 70.16% (248) | hypothetical protein MAB_2404 |
| M. marinum M | MMAR_2705 | - | 1e-119 | 86.85% (251) | putative regulatory protein |
| M. avium 104 | MAV_2887 | - | 1e-117 | 86.00% (250) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_3646 | - | 1e-102 | 78.57% (252) | transcriptional regulator, MerR family protein |
| M. thermoresistible (build 8) | TH_1127 | - | 1e-105 | 80.16% (247) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3047 | - | 1e-118 | 86.45% (251) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3134 | - | 1e-109 | 81.93% (249) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3401|M.gilvum_PYR-GCK MTQPDTPGPSGMSIGVVLDLLRADFPDVTISKIRFLEAEGLVTPERTASG
Mvan_3134|M.vanbaalenii_PYR-1 MTQPDTPALNGMSIGVVLDLLRGDFPDVTISKIRFLEAEGLVTPERTASG
MSMEG_3646|M.smegmatis_MC2_155 MTHPDRSAPAGMSIGAVLDLLRSDFPDVTISKIRFLEAEGLVTPERTASG
Rv1828|M.tuberculosis_H37Rv MSAPDSPALAGMSIGAVLDLLRPDFPDVTISKIRFLEAEGLVTPRRASSG
Mb1859|M.bovis_AF2122/97 MSAPDSPALAGMSIGAVLDLLRPDFPDVTISKIRFLEAEGLVTPRRASSG
MLBr_02075|M.leprae_Br4923 MSAPDSPALVGMSIGAVLDLLRSDFPDVTISKIRFLEAEGLVTPQRSASG
MMAR_2705|M.marinum_M MSAPDSPALAGMSIGAVLELLRPDFPDVTISKIRFLEAEGLVTPQRAASG
MUL_3047|M.ulcerans_Agy99 MSARDSPALAGMSIGAVLELLRPDFPDVTISKIRFLEAEGLVTPQRAASG
MAV_2887|M.avium_104 MSAPDSSALAGMSIGAVLELLRPDFPDVTISKIRFLEAEGLVTPQRAASG
TH_1127|M.thermoresistible__bu MSAPDTPALTGMSIGAVLDLLRPEFPDVTISKIRFLESEGLVTPMRTASG
MAB_2404|M.abscessus_ATCC_1997 MTAPDRPALTGMSIGSVLDLLRPEFPDVTISKIRFLESEGLVTPSRSASG
*: * .. ***** **:*** :*************:****** *::**
Mflv_3401|M.gilvum_PYR-GCK YRRFTAYDCARLRFILTAQRDNYLPLKVIKSQLDALPDGELPQTVSPYGT
Mvan_3134|M.vanbaalenii_PYR-1 YRRFTAYDCARLRFILTAQRDQYLPLKVIKAQLDAQPDGELPQFGSAYAT
MSMEG_3646|M.smegmatis_MC2_155 YRRFTAYDCARLRFILTAQRDQYLPLKVIKAQLDALPDGELPQTGSAYAV
Rv1828|M.tuberculosis_H37Rv YRRFTAYDCARLRFILTAQRDHYLPLKVIRAQLDAQPDGELPPFGSPYVL
Mb1859|M.bovis_AF2122/97 YRRFTAYDCARLRFILTAQRDHYLPLKVIRAQLDAQPDGELPPFGSPYVL
MLBr_02075|M.leprae_Br4923 YRRFTAYDCARLRFILTAQRDHYLPLKVIRAQLDAQPDGELPSFGSPYVA
MMAR_2705|M.marinum_M YRRFTAYDCARLRFILTAQRDHYLPLKVIRSELDAQPDGQLPAIGSPYGV
MUL_3047|M.ulcerans_Agy99 YRRFTAYDCARLRFILTAQRDHYLPLKVIRSELDAQPDGQLPAIGSPYGV
MAV_2887|M.avium_104 YRRFTAYDCARLRFILTAQRDHYLPLKVIRAQLDAQPDGELPAFNTPYTA
TH_1127|M.thermoresistible__bu YRRFTAYDCARLRFILTAQRDQYLPLKVIKAQLDAMPDGELPELPSVYPV
MAB_2404|M.abscessus_ATCC_1997 YRRFSAYDAERLRFILTAQRDHYLPLKVIKEQLDAQPDGALPDLAGFSGG
****:***. ***********:*******: :*** *** **
Mflv_3401|M.gilvum_PYR-GCK PRLVSVG-------QQTASEPAPAPS---PAQVRLSRDDLLERSGVSDDL
Mvan_3134|M.vanbaalenii_PYR-1 PRLVSVG-------GETADVTDVPAG---PTQVRLSREDLLERSEVGGEL
MSMEG_3646|M.smegmatis_MC2_155 PRLVPVGDGGADGGTTTADGVSRAASGMAPTQIRLTREDLLQRSGVDETL
Rv1828|M.tuberculosis_H37Rv PRLVPVAGDSAG----GVGSDTASVS---LTGIRLSREDLLERSEVADEL
Mb1859|M.bovis_AF2122/97 PRLVPVAGDSAG----GVGSDTASVS---LTGIRLSREDLLERSEVADEL
MLBr_02075|M.leprae_Br4923 PRLFSVTGGPGAGVGSGVGSDIPAVS---PAGVRLSREDLLDRSGVADDL
MMAR_2705|M.marinum_M PKLVSVAGNSAGTAGPDGGSDTGAVA---PARVRLSREDLIERSGVDDEL
MUL_3047|M.ulcerans_Agy99 PKLVSVAGNSAGTAGPDGGSDTGAVA---PARVRLSREDLIERSGVDDEL
MAV_2887|M.avium_104 PRLVSVAGTDAG-ANAGLGSDTSAVA---PAHVRLSREELLRRAGVDDEL
TH_1127|M.thermoresistible__bu PRLVSADG--------ASGANAAGVA---PTQVRLSREDLLARSGVDESL
MAB_2404|M.abscessus_ATCC_1997 PRLFAITDG-----DDTNGVARQGFP--PSRPTRLSREDLLSRSGADEVL
*:*.. . **:*::*: *: . *
Mflv_3401|M.gilvum_PYR-GCK LTSLIRAGVITPIFKGAGTTLFDEHSVVIAQCARALADYGVEPRHLRAFR
Mvan_3134|M.vanbaalenii_PYR-1 LNALIKAGIIT---TGPGG-FFDEHSVVIAQCARALADYGVEPRHLRAFR
MSMEG_3646|M.smegmatis_MC2_155 LGALIKNGVITP---GKGG-LFDEHSVVIAQCAKALEEYGVEPRHLRAFR
Rv1828|M.tuberculosis_H37Rv LTALLKAGVITT---GPGG-FFDEHAVVILQCARALAEYGVEPRHLRAFR
Mb1859|M.bovis_AF2122/97 LTALLKAGVITT---GPGG-FFDEHAVVILQCARALAEYGVEPRHLRAFR
MLBr_02075|M.leprae_Br4923 LTALLKAGVITT---GPGG-FFDEHAIVILQCARALSEYGVEPRHLRAFR
MMAR_2705|M.marinum_M LSAVLKAGVITT---GPGG-FFDEHAVVILQCARALSEYGVEPRHLRAFR
MUL_3047|M.ulcerans_Agy99 LSAVLKAGVITT---GPGG-FFDEHAVVILQCARALSEYGVEPRHLRAFR
MAV_2887|M.avium_104 LTALLKAGIITT---GPAG-FFDEHAVVILQCARALSDYGVEPRHLRAFR
TH_1127|M.thermoresistible__bu LSGLVKAGVIKP---GPGG-FFDEHSVVIAQCARALAEYGVEPRHLRAFR
MAB_2404|M.abscessus_ATCC_1997 LTSLIKAGIITS---GPGG-FFDEYTVLIVQCAAELADYGVEPRHLRTFR
* .::: *:*. * . :***::::* *** * :*********:**
Mflv_3401|M.gilvum_PYR-GCK SAADRQSDLIAQIAGPVGKGGKAGARDRADDLAREVAALAITLHTSLIKS
Mvan_3134|M.vanbaalenii_PYR-1 SAADRQSDLIAQIAGPVVKGGKAGARDRADDLAREVAALAITLHTSLIKS
MSMEG_3646|M.smegmatis_MC2_155 SAADRQSDLIAQIAGPVVKAGKAGARDRADDLAREVAALAITLHTSLIKS
Rv1828|M.tuberculosis_H37Rv SAADRQSDLIAQIAGPLVKAGKAGARDRADDLAREVAALAITLHTSLIKS
Mb1859|M.bovis_AF2122/97 SAADRQSDLIAQIAGPLVKAGKAGARDRADDLAREVAALAITLHTSLIKS
MLBr_02075|M.leprae_Br4923 SAADRQSDLIVQIAGPIVKAGKAGARDRADDLAREVAALAITLHTSLIKS
MMAR_2705|M.marinum_M SAADRQSDLIAQIAGPLVKADKAGARDRADDLAREVAALAITLHTSLIKS
MUL_3047|M.ulcerans_Agy99 SAADRQSDLIAQIAGPLVKADKAGARDRADDLAREVAALAITLHTSLIKS
MAV_2887|M.avium_104 SAADRQSDLIAQIAGPVGKAGKAGARDRADDLAREVAALAITLHTSLIKS
TH_1127|M.thermoresistible__bu SAADRQSDLIAQIAGPLVKTDKAGARDRADDLAREVAALAITLHTALIKS
MAB_2404|M.abscessus_ATCC_1997 SAVDRETDLIAQIVGPTVKANKAGARDRADDLIREVAALSIALHGAMIKS
**.**::***.**.** * .*********** ******:*:** ::***
Mflv_3401|M.gilvum_PYR-GCK AVRDVLDR
Mvan_3134|M.vanbaalenii_PYR-1 AVRDVLDR
MSMEG_3646|M.smegmatis_MC2_155 AVRDVLDR
Rv1828|M.tuberculosis_H37Rv AVRDVLHR
Mb1859|M.bovis_AF2122/97 AVRDVLHR
MLBr_02075|M.leprae_Br4923 AVRGVLHR
MMAR_2705|M.marinum_M AVRDVLHR
MUL_3047|M.ulcerans_Agy99 AVRDVLHR
MAV_2887|M.avium_104 AVRDVLDR
TH_1127|M.thermoresistible__bu AVRDVLDR
MAB_2404|M.abscessus_ATCC_1997 AVRGVLDR
***.**.*