For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLQELWFGVIAALFLGFFILEGFDFGVGMLMAPFAHVGMGDPETHRRTALNTIGPVWDGNEVWLITAGAA IFAAFPGWYATVFSALYLPLLAILFGMILRAVAIEWRGKIDDPKWRTGADFGIAAGSWLPALLWGVAFAI LVRGLPVDANGHVALSIPDVLNAYTLLGGLATAGLFSLYGAVFIALKTSGPIRDDAYRFAVWLSLPVAGL VAGFGLWTQLAYGKDWTWLVLAVAGCAQAAATVLVWRRVSDGWAFMCTLIVVAAVVVLLFGALYPNLVPS TLNPQWSLTIHNASSTPYTLKIMTWVTAFFAPLTVAYQTWTYWVFRQRISAERIPPPTGLARRAP*
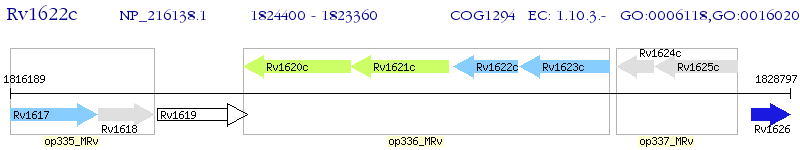
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1622c | cydB | - | 100% (346) | Probable integral membrane cytochrome D ubiquinol oxidase (subunit II) cydB (Cytochrome BD-I oxidase subunit II) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1648c | cydB | 0.0 | 100.00% (346) | integral membrane cytochrome D ubiquinol oxidase (subunit |
| M. gilvum PYR-GCK | Mflv_3586 | - | 1e-140 | 66.96% (342) | cytochrome d ubiquinol oxidase, subunit II |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2631 | - | 1e-125 | 63.74% (342) | cytochrome D ubiquinol oxydase CydB |
| M. marinum M | MMAR_2425 | cydB | 1e-176 | 85.96% (342) | integral membrane cytochrome D ubiquinol oxidase (subunit |
| M. avium 104 | MAV_3165 | cydB | 1e-172 | 83.33% (342) | cytochrome D ubiquinol oxidase, subunit II |
| M. smegmatis MC2 155 | MSMEG_3232 | cydB | 1e-135 | 68.01% (347) | cytochrome D ubiquinol oxidase, subunit II |
| M. thermoresistible (build 8) | TH_1291 | cydB | 1e-144 | 69.10% (343) | Probable integral membrane cytochrome D ubiquinol oxidase |
| M. ulcerans Agy99 | MUL_1603 | cydB | 1e-173 | 84.30% (344) | integral membrane cytochrome D ubiquinol oxidase (subunit |
| M. vanbaalenii PYR-1 | Mvan_2829 | - | 1e-141 | 68.02% (344) | cytochrome d ubiquinol oxidase, subunit II |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1622c|M.tuberculosis_H37Rv ---MVLQELWFGVIAALFLGFFILEGFDFGVGMLMAPFAHVGM---GDPE
Mb1648c|M.bovis_AF2122/97 ---MVLQELWFGVIAALFLGFFILEGFDFGVGMLMAPFAHVGM---GDPE
MMAR_2425|M.marinum_M ---MGLQELWFAVIAVLFLGFFILEGFDFGVGMLMAPFAHFGT---GDPE
MUL_1603|M.ulcerans_Agy99 ---MGLQELWFAVIAVLFLGFFILEGFDFGVGMLMAPFAHFGT---GDPE
MAV_3165|M.avium_104 ---MGLQEVWFGIIGMLFLGFFILEGFDFGVGMLMEPFARVGI---GEPE
Mflv_3586|M.gilvum_PYR-GCK ---MGLQEVWFLLIAVLFLGFLVLEGFDFGVGMLMAPMGTLGP---GDPD
Mvan_2829|M.vanbaalenii_PYR-1 ---MGLQELWFLLIAVLFLGFLVLEGFDFGVGMLMSPMGTIGP---GDPD
TH_1291|M.thermoresistible__bu VISVGLQELWFILIAALFLGFLVLEGFDFGVGMLMGPLGAAGE---GDPE
MSMEG_3232|M.smegmatis_MC2_155 ---MGLQELWFILLAVLFLGFFLLEGFDFGVGMLMSFFGRAAEKRGQDPE
MAB_2631|M.abscessus_ATCC_1997 MNPTALQQFWFVVVAVLFLGFLVLEGFDFGVGMLMHPLGRGDD-------
**:.** ::. *****::************ :.
Rv1622c|M.tuberculosis_H37Rv THRRTALNTIGPVWDGNEVWLITAGAAIFAAFPGWYATVFSALYLPLLAI
Mb1648c|M.bovis_AF2122/97 THRRTALNTIGPVWDGNEVWLITAGAAIFAAFPGWYATVFSALYLPLLAI
MMAR_2425|M.marinum_M IHRRTALNTIGPVWDGNEVWLITGGAAMFAAFPGWYATVFSTLYLPLLAI
MUL_1603|M.ulcerans_Agy99 IHRRTALNTIGPVWDGNEVWLITSGAAMFAAFPGWYATVFSTLYLPLLAI
MAV_3165|M.avium_104 PLRRTALNTIGPVWDGNEVWLIVGGAAMFAAFPGWYATVFSTLYLPLLAI
Mflv_3586|M.gilvum_PYR-GCK NRRRAVLNTIGPVWDANEVWLITAGAAMFAAFPVWYATLFSALYLPLLAI
Mvan_2829|M.vanbaalenii_PYR-1 DRRRAVLNTIGPVWDANEVWLITAGAAMFAAFPNWYATLFSALYLPLLAI
TH_1291|M.thermoresistible__bu PRRRAVLNTIGPVWDANEVWLITAGASMFAAFPNWYATLFSALYLPLLAI
MSMEG_3232|M.smegmatis_MC2_155 PYRRAALNTIGPVWDGNEVWLITAGGAMFAAFPEMYASMFSGLYLALLVI
MAB_2631|M.abscessus_ATCC_1997 RRRRAVLNTIGPVWDGNEVWLITAGGAMFAAFPHWYATVFSGLYLPLLLI
**:.*********.******..*.::***** **::** ***.** *
Rv1622c|M.tuberculosis_H37Rv LFGMILRAVAIEWRGKIDDPKWRTGADFGIAAGSWLPALLWGVAFAILVR
Mb1648c|M.bovis_AF2122/97 LFGMILRAVAIEWRGKIDDPKWRTGADFGIAAGSWLPALLWGVAFAILVR
MMAR_2425|M.marinum_M LFGMIVRSVAIEWRGKVDDPKWRALADFGIAAGSWLPAILWGVAFAILVR
MUL_1603|M.ulcerans_Agy99 LFGMIVRSVAIEWRGKVDDSKWRALADFGIAAGSWLPAILWGVAFAILVR
MAV_3165|M.avium_104 LFGMILRAVAIEWRGKIDDTKWRGWADFGIAAGSWLPAVLWGVAFAVLVR
Mflv_3586|M.gilvum_PYR-GCK LFGMILRIVGIEWRGKINDPQWRRWADYGIAAGSWLPAVLWGVAFAILLR
Mvan_2829|M.vanbaalenii_PYR-1 LFGMILRIVGIEWRGKINDPQWRRWADIGIALGSWLPAVLWGVAFAILLR
TH_1291|M.thermoresistible__bu LFGMIVRIVGIEWRGKIDDERWRKWADAGIAVGSWLPAVLWGVAFAILLR
MSMEG_3232|M.smegmatis_MC2_155 LCAMILRIVAIEWRGKIDDPGWRRWADTGIAVGSWVPAILWGVAFASLVR
MAB_2631|M.abscessus_ATCC_1997 LVSMIVRIVAIEWRGKIDDPRWRARCDLGIAIGSWIPALLWGVAFSSLLA
* .**:* *.******::* ** .* *** ***:**:******: *:
Rv1622c|M.tuberculosis_H37Rv GLPVDANGHVALS-IPDVLNAYTLLGGLATAGLFSLYGAVFIALKTSGPI
Mb1648c|M.bovis_AF2122/97 GLPVDANGHVALS-IPDVLNAYTLLGGLATAGLFSLYGAVFIALKTSGPI
MMAR_2425|M.marinum_M GLPVDADGHVALS-IGDVLNAYTLLGGLATAGLFLFYGAVFVALKTAGPI
MUL_1603|M.ulcerans_Agy99 GLPVDANGRLALS-IGDVLNAYTLLGGLATAGLFLFYGAVFVALKTAGPI
MAV_3165|M.avium_104 GLPVDADGHIHLS-VTDVLNPYTLLGGLATAGLFLLYGAVFVALKTSGAI
Mflv_3586|M.gilvum_PYR-GCK GLPIDAEGRTDIA-FGDVLSPYTLLGGLATGSLFLFYGSVYLALKTSGVL
Mvan_2829|M.vanbaalenii_PYR-1 GLPIDADGRAHPG-IGDILSAYTLLGGLATASLFLFYGSVYLALRTAGAL
TH_1291|M.thermoresistible__bu GLPIDADGRAHPG-LTDVLNGYTLLGGLATASLFAFYGAVFVALKTAGSV
MSMEG_3232|M.smegmatis_MC2_155 GLPVDADKQIHLSFFGDLLNAYTLLGGLATCALFAFHGAVFVSLKTSGEI
MAB_2631|M.abscessus_ATCC_1997 GLPVDGAKQLTLT-VGDVLRPYVLLGGVVFVGLFALHGALFICLKTSGVV
***:*. : . *:* *.****:. .** ::*::::.*:*:* :
Rv1622c|M.tuberculosis_H37Rv RDDAYRFAVWLSLPVAGLVAGFGLWTQLAYGKDWTWLVLAVAGCAQAAAT
Mb1648c|M.bovis_AF2122/97 RDDAYRFAVWLSLPVAGLVAGFGLWTQLAYGKDWTWLVLAVAGCAQAAAT
MMAR_2425|M.marinum_M RDDAYRFAVVLSLPVTVLVAGFGVWTQLAYGKQWTWALLGVAVVAQLAAV
MUL_1603|M.ulcerans_Agy99 RDDAYRFAVVLSLPVTVLVAGFGVWTQLAYGKQWTWALLGVAVVAQMAAV
MAV_3165|M.avium_104 RDDAYRFAVWLSLPVTGLVAGFGLWTQLGYGKDWTWLVLAVAVVAQVAAV
Mflv_3586|M.gilvum_PYR-GCK REDAFRVGKALSVPVIVLAGGFGLWTQLAYGRPWTWIALAAAVVALLAAV
Mvan_2829|M.vanbaalenii_PYR-1 HDDSFRVARVLSLPVIVLAGGFGLWTQLAYGKPWTWLALAVAVVALLISV
TH_1291|M.thermoresistible__bu RADAFRFAKWLSAPVIVLAGGFGLWTQLAHGTDWTWVALGVAVVALLVAV
MSMEG_3232|M.smegmatis_MC2_155 RTDAFGFARLLALPATALVAAFGVWTQVAHGTGWTWIVLAAAVIAQLVAV
MAB_2631|M.abscessus_ATCC_1997 RDDAVAWARKLAAPVIVGAGGYGLWTQLAYGASWTWIALGTAALS-LVAA
: *: . *: *. ...:*:***:.:* *** *..* : :.
Rv1622c|M.tuberculosis_H37Rv VLVWRRVSDGWAFMCTLIVVAAVVVLLFGALYPNLVPSTLNPQWSLTIHN
Mb1648c|M.bovis_AF2122/97 VLVWRRVSDGWAFMCTLIVVAAVVVLLFGALYPNLVPSTLNPQWSLTIHN
MMAR_2425|M.marinum_M LLVWRRVSDGWAFLCIAVVVAAVVLLLFGALYPNLVPSTLNERWNVTIYN
MUL_1603|M.ulcerans_Agy99 LLVWRRVSDGWAFLCIAVVVAAVVLLLFGALYPNLVPSTLNERWNVTVYN
MAV_3165|M.avium_104 LLVWRRASDGWAFTCLALVVAAVVILLFGALYPNLVPSTLDSRWSLTIYN
Mflv_3586|M.gilvum_PYR-GCK ALMWGQTREGIAFVSMVVVIAAVAVLIFGSMYPNLLPSTLNPEWSVTIYN
Mvan_2829|M.vanbaalenii_PYR-1 ALVWKGFREGVAFVSMVVVIAAVAVLIFGSMYPNLLPSTLNPEWSVTIYN
TH_1291|M.thermoresistible__bu GLMWAGRREGWAFFNTVVVITAVTVLIFGAMYPNLLPSTLNPEWSVTIYN
MSMEG_3232|M.smegmatis_MC2_155 AQVFRGSGEGWAFGATSVVVAAVVVLLFGSLFPDLIPSTLNPDWSLTIYN
MAB_2631|M.abscessus_ATCC_1997 ALASRISRDGWAFACTCLTVVAVVALLFGSLYPNLIVSNLDPAYNLTIVN
:* ** :.:.**. *:**:::*:*: *.*: :.:*: *
Rv1622c|M.tuberculosis_H37Rv ASSTPYTLKIMTWVTAFFAPLTVAYQTWTYWVFRQRISAERIPPPTGLAR
Mb1648c|M.bovis_AF2122/97 ASSTPYTLKIMTWVTAFFAPLTVAYQTWTYWVFRQRISAERIPPPTGLAR
MMAR_2425|M.marinum_M ASSTPYTLKIMTWVTALFAPLTVVYQAWTYWVFRQRISAERIPPSIGLAR
MUL_1603|M.ulcerans_Agy99 ASSTPYTLKIMTWVTALFAPLTVVYQAWTYWVFRQRISAERIPPSIGLWL
MAV_3165|M.avium_104 ASSTHYTLKVMTWVTAIMAPVTVVYQAWTYWVFRQRLSADRIPAPVGLTR
Mflv_3586|M.gilvum_PYR-GCK GSSTPYTLRIMTWASLALLPLVLGYQAWSYWVFRKRVTAESIPESIGLTR
Mvan_2829|M.vanbaalenii_PYR-1 GSSTPYTLRIMTWASLTLLPLVLAYQAWSYWVFRKRITAESIPASIGLSR
TH_1291|M.thermoresistible__bu GSSTPYTLQIMSWAALTLLPLVLGYQAWTYWVFRKRISTDAIPPPIGLPR
MSMEG_3232|M.smegmatis_MC2_155 GSSSPYTLKIMTWAALVFAPLVVVYQGWTYWVFSKRISADRIPAPIGLSR
MAB_2631|M.abscessus_ATCC_1997 ASSSPYTLKVMSWAAAITAPVVLVYQGWTYWVFRQRISAEQIPDPVGLTV
.**: ***::*:*.: *:.: ** *:**** :*:::: ** . **
Rv1622c|M.tuberculosis_H37Rv RAP-----------------------------------------------
Mb1648c|M.bovis_AF2122/97 RAP-----------------------------------------------
MMAR_2425|M.marinum_M RPS-----------------------------------------------
MUL_1603|M.ulcerans_Agy99 GAPPDRKGLDHPRAAGPAAVAGVGRVAVLSDRLGDLRSGHLGLCHCVGDR
MAV_3165|M.avium_104 RPS-----------------------------------------------
Mflv_3586|M.gilvum_PYR-GCK RVS-----------------------------------------------
Mvan_2829|M.vanbaalenii_PYR-1 RPS-----------------------------------------------
TH_1291|M.thermoresistible__bu --------------------------------------------------
MSMEG_3232|M.smegmatis_MC2_155 RSS-----------------------------------------------
MAB_2631|M.abscessus_ATCC_1997 R-------------------------------------------------
Rv1622c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1648c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2425|M.marinum_M --------------------------------------------------
MUL_1603|M.ulcerans_Agy99 AGEHGRARRQRPLGTVSTLLAGLAVDVVGAVGVACGDALAAGAPGPARGQ
MAV_3165|M.avium_104 --------------------------------------------------
Mflv_3586|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2829|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1291|M.thermoresistible__bu --------------------------------------------------
MSMEG_3232|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2631|M.abscessus_ATCC_1997 --------------------------------------------------
Rv1622c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1648c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2425|M.marinum_M --------------------------------------------------
MUL_1603|M.ulcerans_Agy99 RGDRRAQWAGAGVGDGPPAQRIGGTTRCRRGGGNPRTGRLATLFHRLPAH
MAV_3165|M.avium_104 --------------------------------------------------
Mflv_3586|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2829|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1291|M.thermoresistible__bu --------------------------------------------------
MSMEG_3232|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2631|M.abscessus_ATCC_1997 --------------------------------------------------
Rv1622c|M.tuberculosis_H37Rv ---------------------------------------------
Mb1648c|M.bovis_AF2122/97 ---------------------------------------------
MMAR_2425|M.marinum_M ---------------------------------------------
MUL_1603|M.ulcerans_Agy99 AVSCGDPDPGHRGDDRGLRREVGADRGDHPSTDPGLHGLDRVGYR
MAV_3165|M.avium_104 ---------------------------------------------
Mflv_3586|M.gilvum_PYR-GCK ---------------------------------------------
Mvan_2829|M.vanbaalenii_PYR-1 ---------------------------------------------
TH_1291|M.thermoresistible__bu ---------------------------------------------
MSMEG_3232|M.smegmatis_MC2_155 ---------------------------------------------
MAB_2631|M.abscessus_ATCC_1997 ---------------------------------------------