For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGLRPARVVRPARSGMLKGVTDPLQHGAFEPGWQSAPPGYPPPYPQYPGPGSYFDPFAPYGRHPVTGQPF SDKSKTVAGLLQLLGLFGIAGIGRIYLGHTGLGIAQLLVGWVTCGLGAVIWGVIDALLILTDKVGDPWGR PLRDGS
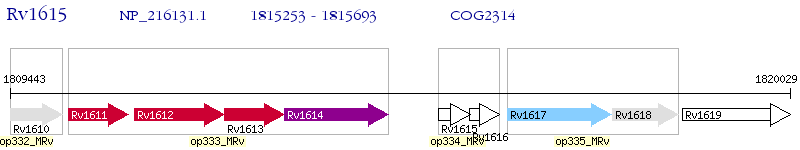
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1615 | - | - | 100% (146) | Probable hypothetical membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1641 | - | 3e-87 | 100.00% (146) | hypothetical protein Mb1641 |
| M. gilvum PYR-GCK | Mflv_3595 | - | 2e-41 | 59.44% (143) | TM2 domain-containing protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2641c | - | 2e-34 | 62.07% (116) | hypothetical protein MAB_2641c |
| M. marinum M | MMAR_2417 | - | 4e-55 | 79.51% (122) | hypothetical protein MMAR_2417 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3223 | - | 5e-41 | 78.72% (94) | TM2 domain-containing protein |
| M. thermoresistible (build 8) | TH_1095 | - | 3e-40 | 59.26% (135) | Probable hypothetical membrane protein |
| M. ulcerans Agy99 | MUL_1595 | - | 3e-55 | 79.51% (122) | hypothetical protein MUL_1595 |
| M. vanbaalenii PYR-1 | Mvan_2820 | - | 2e-45 | 66.93% (127) | TM2 domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3595|M.gilvum_PYR-GCK -------MTEPPFSGNEGADKYPPPPQ--QPGYVPPGSQYVPPPPGAYPP
Mvan_2820|M.vanbaalenii_PYR-1 -------MTEPPFSGNEGGPSYTPPPP--QPGYVPPGPQYAPPPPGAYPP
Rv1615|M.tuberculosis_H37Rv MGLRPARVVRPARSGMLKGVTDPLQHGAFEPGWQSAPPGYPPP-YPQYPG
Mb1641|M.bovis_AF2122/97 MGLRPARVVRPARSGMLKGVTDPLQHGAFEPGWQSAPPGYPPP-YPQYPG
MMAR_2417|M.marinum_M ------MVTDQSWAG---GGAGPGQQ----PGY---PPPFPPPYYPDYSA
MUL_1595|M.ulcerans_Agy99 -------MTDQSWAG---GGAGPGQQ----PGY---PPPFPPPYYPDYSA
MSMEG_3223|M.smegmatis_MC2_155 --------------------------------------------------
TH_1095|M.thermoresistible__bu -------MTESDKPGN-SGADVPPPSAGSPPPYSSPAGQYPPP----PPG
MAB_2641c|M.abscessus_ATCC_199 -------MTES----------TPPPFG--------TPPPVPPAPGVPYPP
Mflv_3595|M.gilvum_PYR-GCK SGAYPPPVQGGYPGGYPGSYVDPMAPYGRHPLTGEPLSDKSKTVAGLLQL
Mvan_2820|M.vanbaalenii_PYR-1 P-----------AGGYAGGYVDPAAPWGRHPVTGEPFSEKSKVVGGLLQL
Rv1615|M.tuberculosis_H37Rv PG----------------SYFDPFAPYGRHPVTGQPFSDKSKTVAGLLQL
Mb1641|M.bovis_AF2122/97 PG----------------SYFDPFAPYGRHPVTGQPFSDKSKTVAGLLQL
MMAR_2417|M.marinum_M PR----------------SYYDPAAPFGRHPVTGQPFSDKSKTVAGLLQL
MUL_1595|M.ulcerans_Agy99 PR----------------SYYDPAAPFGRHPVTGQPFSDKSKTVAGLLQL
MSMEG_3223|M.smegmatis_MC2_155 ------------------MYGDPTAPYGRHPVTGEPFSDKSKIVAGLLQL
TH_1095|M.thermoresistible__bu PY----------------PYADMSAPFGRHPVTGEPLSDKSKIAAGLLQL
MAB_2641c|M.abscessus_ATCC_199 P------------------YVDPAAPYGRDPATGAPYSDKSKLVAGLLQL
* * **:**.* ** * *:*** ..*****
Mflv_3595|M.gilvum_PYR-GCK LGIFGFLGIGRMYIGQVGFGVAQLVGCLVLGAVTCGVGFIVPVIWGIVDA
Mvan_2820|M.vanbaalenii_PYR-1 LGLFGLVGIGRIYLGYTSLGVIQLV----VGLITCGIG---AIIWGIVDA
Rv1615|M.tuberculosis_H37Rv LGLFGIAGIGRIYLGHTGLGIAQLL----VGWVTCGLG---AVIWGVIDA
Mb1641|M.bovis_AF2122/97 LGLFGIAGIGRIYLGHTGLGIAQLL----VGWVTCGLG---AVIWGVIDA
MMAR_2417|M.marinum_M LGLFGIAGIGRIYMGHTGMGIAQLL----VGWLTCGLG---ALVWGVIDA
MUL_1595|M.ulcerans_Agy99 LGLFGIAGIGRIYMGHTGMGIAQLL----VGWLTCGLG---ALVWGVIDA
MSMEG_3223|M.smegmatis_MC2_155 IGLVGFVGFGRIYLGQTGLGIAQLI----VGLVTCGIG---AFIWGIVDA
TH_1095|M.thermoresistible__bu VGLLGVVGIGRMYIGHTGLGIAQLV----IGILTCGIG---AVVWGIVDA
MAB_2641c|M.abscessus_ATCC_199 LGLVGFLGFGRIYLGQTLLGVVQLL----IGFVTFGIV---AVIWAIVDA
:*:.*. *:**:*:* . :*: **: :* :* *: ..:*.::**
Mflv_3595|M.gilvum_PYR-GCK VLMFTDKVRDPQGRPLRDGT
Mvan_2820|M.vanbaalenii_PYR-1 ILILTDKVRDPEGRPLRDGT
Rv1615|M.tuberculosis_H37Rv LLILTDKVGDPWGRPLRDGS
Mb1641|M.bovis_AF2122/97 LLILTDKVGDPWGRPLRDGS
MMAR_2417|M.marinum_M VLILTDKVSDPWGRPLRDGT
MUL_1595|M.ulcerans_Agy99 VLILTDKVSDPWGRPLRDGT
MSMEG_3223|M.smegmatis_MC2_155 ILILTDKVRDPYGRPLRDGT
TH_1095|M.thermoresistible__bu VLILTDKVRDSEGRRLRDDL
MAB_2641c|M.abscessus_ATCC_199 VLIFGGKVTDKVGRQLRD--
:*:: .** * ** ***