For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKPFAPRRLYTPRTSRTLAPRLDPEAVGRTTESIARFFGTGRYLLVQTLLVLTWIVLNLFAVGLRWDPYP FILLNLAFSTQASYAAPLILLAQNRQEKRDRAVFEEDRRRAAQTKADTEYNARELAALRLAIGEVPTRDY LRHELDSLRALLAELQPTDPDVAQPRVADEAEQHAKKSG*
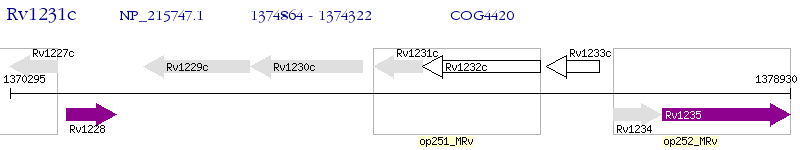
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1231c | - | - | 100% (180) | PROBABLE MEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1263c | - | 2e-99 | 100.00% (180) | hypothetical protein Mb1263c |
| M. gilvum PYR-GCK | Mflv_2207 | - | 1e-59 | 64.25% (179) | hypothetical protein Mflv_2207 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1523c | - | 5e-60 | 73.25% (157) | hypothetical protein MAB_1523c |
| M. marinum M | MMAR_4210 | - | 4e-74 | 79.89% (179) | hypothetical protein MMAR_4210 |
| M. avium 104 | MAV_1370 | - | 2e-72 | 76.54% (179) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_5066 | - | 3e-60 | 64.84% (182) | integral membrane protein |
| M. thermoresistible (build 8) | TH_2064 | - | 5e-56 | 70.44% (159) | PROBABLE MEMBRANE PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4489 | - | 7e-61 | 67.82% (174) | hypothetical protein Mvan_4489 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2207|M.gilvum_PYR-GCK MSEPSSRQRLDTPRVSRSFAP--RLDIEAVGRFSEGIARFLGTGRYLAIQ
Mvan_4489|M.vanbaalenii_PYR-1 MSDASSRQRLDTPRVSRGLAP--RLDIEAVGRFSERIARFLGTGRYLAIQ
MSMEG_5066|M.smegmatis_MC2_155 MSEPTSRQRLDTPRVWRGPRL--QVDVEAVGRVSESIARFLGTGRYLAIQ
TH_2064|M.thermoresistible__bu VSELSARQRLDTPRASRGLDLGLGLDVDAMGRFSERIARFLGTGRYLAAQ
MAB_1523c|M.abscessus_ATCC_199 MSDASARQRLDTPRTSRRLSL--GLDVEAVGRVSENIARFLGTGRYLAIQ
Rv1231c|M.tuberculosis_H37Rv MSKPFAPRRLYTPRTSRTLAP--RLDPEAVGRTTESIARFFGTGRYLLVQ
Mb1263c|M.bovis_AF2122/97 MSKPFAPRRLYTPRTSRTLAP--RLDPEAVGRTTESIARFFGTGRYLLVQ
MMAR_4210|M.marinum_M MSKTPAARRLYTPRTSRAFAP--RLDPEAVGRTTESIARFFGTGRYLLVQ
MAV_1370|M.avium_104 MSKSTAVRRLYTPRTSRRYSP--RLDPETVGQITESIARFFGTGRYLLLQ
:*. : :** ***. * :* :::*: :* ****:****** *
Mflv_2207|M.gilvum_PYR-GCK TIVVIVWILLNLGAFAWQWDPYPFILLNLAFSTQAAYAAPLILLAQNRQE
Mvan_4489|M.vanbaalenii_PYR-1 TILVIVWVLLNLGAFAWQWDPYPFILLNLAFSTQAAYAAPLILLAQNRQE
MSMEG_5066|M.smegmatis_MC2_155 TIFVVVWIALNLFAVGYEWDPYPFILLNLAFSTQAAYAAPLILLAQNRQE
TH_2064|M.thermoresistible__bu TIVVVVWIALNIGAFAWQWDPYPFILLNLAFSTQAAYAAPLILLAQNRQE
MAB_1523c|M.abscessus_ATCC_199 TIFVVVWIALNLFAVGLEWDPYPFILLNLAFSTQAAYAAPLILLAQNRQE
Rv1231c|M.tuberculosis_H37Rv TLLVLTWIVLNLFAVGLRWDPYPFILLNLAFSTQASYAAPLILLAQNRQE
Mb1263c|M.bovis_AF2122/97 TLLVLTWIVLNLFAVGLRWDPYPFILLNLAFSTQASYAAPLILLAQNRQE
MMAR_4210|M.marinum_M TLIVVIWISLNLFAVKLRWDPYPFILLNLAFSTQASYAAPLILLAQNRQE
MAV_1370|M.avium_104 TIVVAVWIVVNVFAVRLRWDPYPFILLNLAFSTQAAYAAPLILLAQNRQE
*:.* *: :*: *. .*****************:**************
Mflv_2207|M.gilvum_PYR-GCK NRDRVALDEDRRRAQQTKADTEFLARELAALRLAVGEVATRDYLRRELDE
Mvan_4489|M.vanbaalenii_PYR-1 NRDRVALEEDRRRAEQTKADTEFLARELAALRLAVGEVATRDYLRRELEE
MSMEG_5066|M.smegmatis_MC2_155 NRDRVSLEEDRRRAEQTKADTEFLARELASLRLAIGEVATRDYLRRELEE
TH_2064|M.thermoresistible__bu NRDRVALEEDRRRAEQTKADTEYLARELASLRLAIGEVATRDYLRRELDE
MAB_1523c|M.abscessus_ATCC_199 NRDRVALEEDRRRAEQTKADTEYLARELAALRLAVGEVATRDYLRRELEQ
Rv1231c|M.tuberculosis_H37Rv KRDRAVFEEDRRRAAQTKADTEYNARELAALRLAIGEVPTRDYLRHELDS
Mb1263c|M.bovis_AF2122/97 KRDRAVFEEDRRRAAQTKADTEYNARELAALRLAIGEVPTRDYLRHELDS
MMAR_4210|M.marinum_M NRDRVLLDGDRRRAAQTKADTEYLARELAALRLAIGAVPTRDYLRHELES
MAV_1370|M.avium_104 NRDRVALEEDRRRAAQTKADTEYLARELAAVRLAVGEVVTREYLRHELDD
:***. :: ***** *******: *****::***:* * **:***:**:.
Mflv_2207|M.gilvum_PYR-GCK IREMIEKLSGEK-DAEPGRAGKQDTGARKTKKSS
Mvan_4489|M.vanbaalenii_PYR-1 IRELIQELHSRD-RAETGGASKRDAGERKSKKSG
MSMEG_5066|M.smegmatis_MC2_155 IRDMISELQQPPPTTDRAGSGKGDGGERRSKRGG
TH_2064|M.thermoresistible__bu LREMLSELQRHGGTGDTGDFGPPEPSGRPAPPSE
MAB_1523c|M.abscessus_ATCC_199 LHEALESIREKNLL--------------------
Rv1231c|M.tuberculosis_H37Rv LRALLAELQPTD--PDVAQPRVADEAEQHAKKSG
Mb1263c|M.bovis_AF2122/97 LRALLAELQPTD--PDVAQPRVADEAEQHAKKSG
MMAR_4210|M.marinum_M LRDLVAELQSDQ--PDTET----DGTERRAKKTH
MAV_1370|M.avium_104 LRELLAELRPQS--ADGEPVSGADNREQTAKKSR
:: : .: