For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADPGSVGHVFRRAFSWLPAQFASQSDAPVGAPRQFRSTEHLSIEAIAAFVDGELRMNAHLRAAHHLSLC AQCAAEVDDQSRARAALRDSHPIRIPSTLLGLLSEIPRCPPEGPSKGSSGGSSQGPPDGAAAGFGDRFAD GDGGNRGRQSRVRR
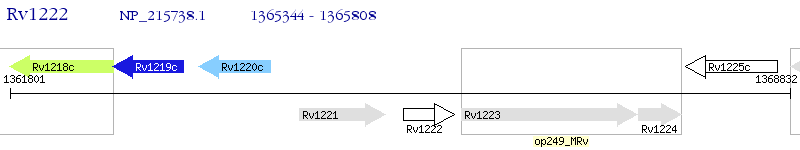
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1222 | - | - | 100% (154) | hypothetical protein Rv1222 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1254 | - | 1e-87 | 100.00% (154) | hypothetical protein Mb1254 |
| M. gilvum PYR-GCK | Mflv_2201 | - | 4e-39 | 68.60% (121) | hypothetical protein Mflv_2201 |
| M. leprae Br4923 | MLBr_01077 | - | 1e-52 | 71.13% (142) | hypothetical protein MLBr_01077 |
| M. abscessus ATCC 19977 | MAB_1363 | - | 2e-38 | 62.02% (129) | hypothetical protein MAB_1363 |
| M. marinum M | MMAR_4215 | - | 7e-52 | 84.07% (113) | hypothetical protein MMAR_4215 |
| M. avium 104 | MAV_1365 | - | 1e-50 | 86.24% (109) | RNA polymerase sigma-70 factor |
| M. smegmatis MC2 155 | MSMEG_5071 | - | 8e-47 | 80.36% (112) | hypothetical protein MSMEG_5071 |
| M. thermoresistible (build 8) | TH_0544 | - | 1e-43 | 72.88% (118) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4518 | - | 5e-52 | 84.07% (113) | hypothetical protein MUL_4518 |
| M. vanbaalenii PYR-1 | Mvan_4495 | - | 2e-41 | 69.75% (119) | hypothetical protein Mvan_4495 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2201|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4495|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1222|M.tuberculosis_H37Rv --------------------------------------------------
Mb1254|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01077|M.leprae_Br4923 --------------------------------------------------
MMAR_4215|M.marinum_M --------------------------------------------------
MUL_4518|M.ulcerans_Agy99 --------------------------------------------------
MAV_1365|M.avium_104 MDRGARETGNTEWQLPVAANDEMPLIGMPNSEELIITTLLSPSSMSHAHD
TH_0544|M.thermoresistible__bu --------------------------------------------------
MSMEG_5071|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1363|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2201|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4495|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1222|M.tuberculosis_H37Rv --------------------------------------------------
Mb1254|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01077|M.leprae_Br4923 --------------------------------------------------
MMAR_4215|M.marinum_M --------------------------------------------------
MUL_4518|M.ulcerans_Agy99 --------------------------------------------------
MAV_1365|M.avium_104 PSADGWAEPSDGLQGTAVFDATGDKTAMPSWDELVRQHADRVYRLAYRLS
TH_0544|M.thermoresistible__bu --------------------------------------------------
MSMEG_5071|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1363|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2201|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4495|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1222|M.tuberculosis_H37Rv --------------------------------------------------
Mb1254|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01077|M.leprae_Br4923 --------------------------------------------------
MMAR_4215|M.marinum_M --------------------------------------------------
MUL_4518|M.ulcerans_Agy99 --------------------------------------------------
MAV_1365|M.avium_104 GNQHDAEDLTQETFIRVFRSVQNYQPGTFEGWLHRITTNLFLDMVRRRSR
TH_0544|M.thermoresistible__bu --------------------------------------------------
MSMEG_5071|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1363|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2201|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4495|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1222|M.tuberculosis_H37Rv --------------------------------------------------
Mb1254|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01077|M.leprae_Br4923 --------------------------------------------------
MMAR_4215|M.marinum_M --------------------------------------------------
MUL_4518|M.ulcerans_Agy99 --------------------------------------------------
MAV_1365|M.avium_104 IRMEALPEDYERVPADEPNPEEIYHDSRLGPDLQAALDSLPPEFRAAVVL
TH_0544|M.thermoresistible__bu --------------------------------------------------
MSMEG_5071|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1363|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2201|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4495|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1222|M.tuberculosis_H37Rv --------------------------------------------------
Mb1254|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01077|M.leprae_Br4923 --------------------------------------------------
MMAR_4215|M.marinum_M --------------------------------------------------
MUL_4518|M.ulcerans_Agy99 --------------------------------------------------
MAV_1365|M.avium_104 CDIEGLSYEEIGATLGVKLGTVRSRIHRGRQALRDYLAAHPDHDALRASL
TH_0544|M.thermoresistible__bu --------------------------------------------------
MSMEG_5071|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1363|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2201|M.gilvum_PYR-GCK ------------------------------------------MERGAVVF
Mvan_4495|M.vanbaalenii_PYR-1 -----------------------------------------------MVF
Rv1222|M.tuberculosis_H37Rv ------------------------------------------------MA
Mb1254|M.bovis_AF2122/97 ------------------------------------------------MA
MLBr_01077|M.leprae_Br4923 -----------------------------------------------MMA
MMAR_4215|M.marinum_M ------------------------------------------------MA
MUL_4518|M.ulcerans_Agy99 ------------------------------------------------MA
MAV_1365|M.avium_104 GVVSARPANVRAAIQRLCGLGLGIGRPVALHSKKANKAAIRMNGKELAMP
TH_0544|M.thermoresistible__bu ------------------------------------------------MA
MSMEG_5071|M.smegmatis_MC2_155 ------------------------------------------------MA
MAB_1363|M.abscessus_ATCC_1997 ------------------------------------------------MV
:
Mflv_2201|M.gilvum_PYR-GCK DPG---HAFRRAFS------WLP----SQSDSPVG-PRQFGSTEHLSTEA
Mvan_4495|M.vanbaalenii_PYR-1 DPG---HAFRRAFS------WLPSQLASQSDDPVG-PRQFGSTEHLSIEA
Rv1222|M.tuberculosis_H37Rv DPGSVGHVFRRAFS------WLPAQFASQSDAPVGAPRQFRSTEHLSIEA
Mb1254|M.bovis_AF2122/97 DPGSVGHVFRRAFS------WLPAQFASQSDAPVGAPRQFRSTEHLSIEA
MLBr_01077|M.leprae_Br4923 DRG---QVFRRVFS------WLPAQFASQNDAPVGAPRRFGSTEHLSVEA
MMAR_4215|M.marinum_M DRG---HVFRRAFS------WLPAQFASQSDAPVGAPRRFRSTEHLSIEA
MUL_4518|M.ulcerans_Agy99 DRG---HVFRRAFS------WLPAQFASQSDAPVGAPRRFRSTEHLSIEA
MAV_1365|M.avium_104 DRG---HVFRRAFS------WLPAQFASQSDAPVGAPRQFGSTEHLSVEA
TH_0544|M.thermoresistible__bu EPG---HLFRRAFS------WLPAQFAPQNDAPVGAPRQFGSTEHLSTEA
MSMEG_5071|M.smegmatis_MC2_155 DPG---HVFRRAFS------WLPSQFASQSDAPVGAPRQFGSTEHLSVEA
MAB_1363|M.abscessus_ATCC_1997 DSG----SFRRAFSSFSSFSRLPSPFASQSGAPVGAPRQFGSTEHLSTEA
: * ***.** ** .*.. *** **:* ****** **
Mflv_2201|M.gilvum_PYR-GCK IAAFADGELRMTAHLRAAHHLSLCSECAQEVDAQRQAREALRDSCPVDIP
Mvan_4495|M.vanbaalenii_PYR-1 VAAFVDGELRMSAHLRAAHHLSLCPECALEVDAQRQAREALRESRPVAMP
Rv1222|M.tuberculosis_H37Rv IAAFVDGELRMNAHLRAAHHLSLCAQCAAEVDDQSRARAALRDSHPIRIP
Mb1254|M.bovis_AF2122/97 IAAFVDGELRMNAHLRAAHHLSLCAQCAAEVDDQSRARAALRDSHPIRIP
MLBr_01077|M.leprae_Br4923 IAAFVDGELRMNAHLRAAHHISLCAQCAAEVDDQSRTRAALRDSHPIRIP
MMAR_4215|M.marinum_M IAAFVDGELRMNAHLRAAHHLSMCPQCAAEVEDHTRARAALRDSHPIRIP
MUL_4518|M.ulcerans_Agy99 IAAFVDGELRMNAHLRAAHHLSMCPQCAAEVEDHTRARAALRDSHPIRIP
MAV_1365|M.avium_104 IAAFVDGELRMNAHLRAAHHLSQCAQCAAEVDDQSRARAALRDSRPIRIP
TH_0544|M.thermoresistible__bu VAAFVDGELGMTAHLRAAHHLSLCPQCAAEVDAQAQARAALRESRPVDVP
MSMEG_5071|M.smegmatis_MC2_155 IAAFVDGELRMSAHLRAAHHLSLCPECAAEVDAQSQARTALRESCPIAIP
MAB_1363|M.abscessus_ATCC_1997 IAAFVDGELRMSAHLRAAHHLSMCAECALEIDAQRQARTALRDSGAIRVP
:***.**** *.********:* *.:** *:: : ::* ***:* .: :*
Mflv_2201|M.gilvum_PYR-GCK SSLLGLLSQIPNRTP------------VDAQEPPES---------PQLAD
Mvan_4495|M.vanbaalenii_PYR-1 SSLLGLLSQIPNHTP------------VEAPEPADS---------PQLAD
Rv1222|M.tuberculosis_H37Rv STLLGLLSEIPRCPPEGPSKGSSGGSSQGPPDGAAAG------FGDRFAD
Mb1254|M.bovis_AF2122/97 STLLGLLSEIPRCPPEGPSKGSSGGSSQGPPDGAAAG------FGDRFAD
MLBr_01077|M.leprae_Br4923 STLFGLLTAIPRCSPDYTSP-----VSEPFSEGSVS---------DRFVD
MMAR_4215|M.marinum_M TALLGMLSEIPHHP------------SEGAPEDTTG-------LADSFAE
MUL_4518|M.ulcerans_Agy99 TALLGMLSEIPHHP------------SEGAPEDTTG-------LADSFAE
MAV_1365|M.avium_104 ANLLGMLAEIPYESP-------------DAP--------------DRLAD
TH_0544|M.thermoresistible__bu NSLWGLLSEIPNRT------------SATTPEDPKP---------PPFAD
MSMEG_5071|M.smegmatis_MC2_155 NSLLGMLSQIPHRTP------------EVTPDVSEQ---------AKFAD
MAB_1363|M.abscessus_ATCC_1997 GSLLGLLSQIPHIPH-----------DVAPPQPSAHEGELPPALGGDAVR
* *:*: ** . .
Mflv_2201|M.gilvum_PYR-GCK GPPRARRKRR----
Mvan_4495|M.vanbaalenii_PYR-1 GAQRSRRKRR----
Rv1222|M.tuberculosis_H37Rv GDGGNRGRQSRVRR
Mb1254|M.bovis_AF2122/97 GDGGNRGRQSRVRR
MLBr_01077|M.leprae_Br4923 GVAREQGKR-----
MMAR_4215|M.marinum_M RQARDQRKRR----
MUL_4518|M.ulcerans_Agy99 RQARDQRKRR----
MAV_1365|M.avium_104 RDARDRRRRP----
TH_0544|M.thermoresistible__bu WQPRGWRKRR----
MSMEG_5071|M.smegmatis_MC2_155 DPTRGRRKRR----
MAB_1363|M.abscessus_ATCC_1997 PDSRARRKRR----
::