For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YYLLILAVVFERLAELVVAQRNARWSFAQGGKEFGRPHYVVMVILHTALLLGCVVEPWALHRPFIPWLGW PMLAVVVASQGLRWWCVKSLGKRWNTRVIVLPHATLVRRGPYRWMRHPNYVAVVAEGFALPLVHTAWLTA LVFTLANATLLTVRLRVENSVLGYI*
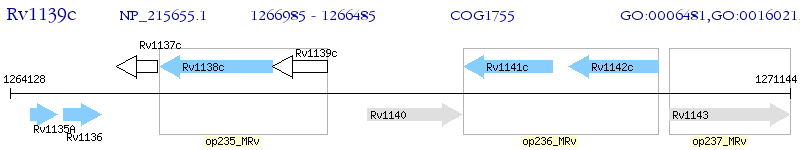
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1139c | - | - | 100% (166) | hypothetical protein Rv1139c |
| M. tuberculosis H37Rv | Rv2437 | - | 1e-07 | 39.19% (74) | hypothetical protein Rv2437 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1171c | - | 9e-97 | 100.00% (166) | hypothetical protein Mb1171c |
| M. gilvum PYR-GCK | Mflv_5141 | - | 4e-60 | 67.28% (162) | isoprenylcysteine carboxyl methyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4314 | - | 6e-83 | 84.15% (164) | hypothetical protein MMAR_4314 |
| M. avium 104 | MAV_1279 | - | 3e-79 | 80.61% (165) | isoprenylcysteine carboxyl methyltransferase (icmt) family |
| M. smegmatis MC2 155 | MSMEG_0809 | - | 2e-52 | 64.78% (159) | isoprenylcysteine carboxyl methyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0980 | - | 6e-82 | 82.93% (164) | hypothetical protein MUL_0980 |
| M. vanbaalenii PYR-1 | Mvan_1195 | - | 4e-58 | 65.43% (162) | isoprenylcysteine carboxyl methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1139c|M.tuberculosis_H37Rv ---MYYLLILAVVFERLAELVVAQRNARWSFAQGGKEFGRPHYVVMVILH
Mb1171c|M.bovis_AF2122/97 ---MYYLLILAVVFERLAELVVAQRNARWSFAQGGKEFGRPHYVVMVILH
MMAR_4314|M.marinum_M MVSPYYLLILAIVVERLAELVIAQRNARWSFAQGGKEFGQPHYVVMVVLH
MUL_0980|M.ulcerans_Agy99 MVSPYYLLILAIVVERLAELVIAQRNARWSFAQGGKEFGQPHYVVMVVLH
MAV_1279|M.avium_104 ---MYYLLVLAVGLERVAELLVSTRNARWSFTQGGKEFGRSHYPVMVFIH
Mflv_5141|M.gilvum_PYR-GCK -MFAYTLLIAAVAVERLAELVVSQRNLKWSKARGGVEFGAGHYPVMVVLH
Mvan_1195|M.vanbaalenii_PYR-1 -MLAYTLLIAAVALERVAELVVSQRNLRWSRARGGVEFGATHYPLMVLLH
MSMEG_0809|M.smegmatis_MC2_155 -MKAFTALVAAVAAERLAELVVSKRNLEWSKTHGGKEFGAGHYPVMVALH
: *: *: **:***::: ** .** ::** *** ** :** :*
Rv1139c|M.tuberculosis_H37Rv TALLLGCVVEPWALHRPFIPWLGWPMLAVVVASQGLRWWCVKSLGKRWNT
Mb1171c|M.bovis_AF2122/97 TALLLGCVVEPWALHRPFIPWLGWPMLAVVVASQGLRWWCVKSLGKRWNT
MMAR_4314|M.marinum_M SLLLVGCVIEPWALHRPFIPWLGWPMVAVLVLSQGLRWWCVTSLGRRWNT
MUL_0980|M.ulcerans_Agy99 SLLLVGCVIEPWALYRPFIPWLGWPMVAVLVLSQGLRWWCVTSLGRRWNT
MAV_1279|M.avium_104 TALLAGCLVEPWALHRPFLGWLGWPMLAVVAASQGLRWWCITTLGRRWNT
Mflv_5141|M.gilvum_PYR-GCK TALLVGCLVE--ASHREFIPALGWTMFAVAIAAQALRWWCITTLGPQWNT
Mvan_1195|M.vanbaalenii_PYR-1 TVLLAACLVE--AAHREFIPVLGWSMFALVVAAQGLRWWCITTLGHQWNT
MSMEG_0809|M.smegmatis_MC2_155 TGLLAGAALE--ARRRRVRPALGRPMFAVVLAAQGLRWWCIKTLGPQWNT
: ** .. :* * * . ** .*.*: :*.*****:.:** :***
Rv1139c|M.tuberculosis_H37Rv RVIVLPHATLVRRGPYRWMRHPNYVAVVAEGFALPLVHTAWLTALVFTLA
Mb1171c|M.bovis_AF2122/97 RVIVLPHATLVRRGPYRWMRHPNYVAVVAEGFALPLVHTAWLTALVFTLA
MMAR_4314|M.marinum_M RVIVLPDEPLVQKGPYRFLHHPNYVAVVAEGFALPLVHTAWVTALSFTVA
MUL_0980|M.ulcerans_Agy99 RVIVLPDEPLAQKGPYRFLHHPNYVAVVAEGFALPLVHTAWVTALSFTVA
MAV_1279|M.avium_104 RVIVLPQAPLVRDGPYRWLHHPNYVAVVAEGLALPLVHTAWLTAAVFTLA
Mflv_5141|M.gilvum_PYR-GCK RVVVVPNAGRVTGGPYRFMSHPNYVAVVVEGIALPLVHTAWITALVFTVL
Mvan_1195|M.vanbaalenii_PYR-1 RVVVVPGAGRVTGGPYRFLSHPNYVAVVVEGIALPLVHTAWVTALAFTLL
MSMEG_0809|M.smegmatis_MC2_155 RVIVVPGATRVLAGPYRFLPHPNYVAVVAEGVALPLVGGAWRTALGFTIA
**:*:* . ****:: ********.**.***** ** ** **:
Rv1139c|M.tuberculosis_H37Rv NATLLTVRLRVENSVLGYI----
Mb1171c|M.bovis_AF2122/97 NATLLTVRLRVENSVLGYI----
MMAR_4314|M.marinum_M NAILLTVRLRVENSVLGYS----
MUL_0980|M.ulcerans_Agy99 NAILLTVRLRVENSVLGYS----
MAV_1279|M.avium_104 NAALLRVRLRVENSALGYT----
Mflv_5141|M.gilvum_PYR-GCK NAALLWTRISVENKALRTLTAAR
Mvan_1195|M.vanbaalenii_PYR-1 NAALLWMRISVEDKALRTLHT--
MSMEG_0809|M.smegmatis_MC2_155 NALLLRTRIRVENDALRSLT---
** ** *: **:..*