For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGVELGSNSEVGALRVVILHRPGAELRRLTPRNTDQLLFDGLPWVSRAQDEHDEFAELLASRGAEVLLLS DLLTEALHHSGAARMQGIAAAVDAPRLGLPLAQELSAYLRSLDPGRLAHVLTAGMTFNELPSDTRTDVSL VLRMHHGGDFVIEPLPNLVFTRDSSIWIGPRVVIPSLALRARVREASLTDLIYAHHPRFTGVRRAYESRT APVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLFDDDLAHTVLAVPIAQQRAQMHLDTVCTMVDTDT MVMYANVVDTLEAFTIQRTPDGVTIGDAAPFAEAAAKAMGIDKLRVIHTGMDPVVAEREQWDDGNNTLAL APGVVVAYERNVQTNARLQDAGIEVLTIAGSELGTGRGGPRCMSCPAARDPL
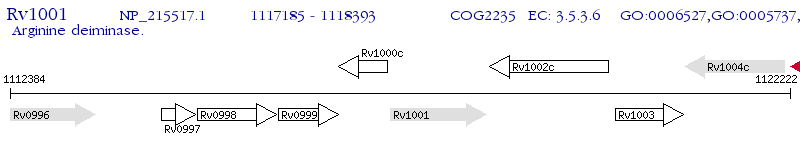
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1001 | arcA | - | 100% (402) | arginine deiminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1028 | arcA | 0.0 | 100.00% (402) | arginine deiminase |
| M. gilvum PYR-GCK | Mflv_1923 | - | 0.0 | 80.65% (398) | arginine deiminase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1121 | - | 1e-173 | 78.20% (399) | arginine deiminase |
| M. marinum M | MMAR_4492 | arcA | 0.0 | 88.47% (399) | arginine deiminase ArcA |
| M. avium 104 | MAV_1125 | arcA | 0.0 | 89.75% (400) | arginine deiminase |
| M. smegmatis MC2 155 | MSMEG_5448 | arcA | 0.0 | 80.75% (400) | arginine deiminase |
| M. thermoresistible (build 8) | TH_0930 | arcA | 2e-92 | 78.24% (216) | PROBABLE ARGININE DEIMINASE ARCA (ADI) (AD) (ARGININE |
| M. ulcerans Agy99 | MUL_4664 | arcA | 0.0 | 87.72% (399) | arginine deiminase |
| M. vanbaalenii PYR-1 | Mvan_4810 | - | 1e-180 | 79.90% (398) | arginine deiminase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1001|M.tuberculosis_H37Rv --MGVELGSNSEVGALRVVILHRPGAELRRLTPRNTDQLLFDGLPWVSRA
Mb1028|M.bovis_AF2122/97 --MGVELGSNSEVGALRVVILHRPGAELRRLTPRNTDQLLFDGLPWVSRA
MMAR_4492|M.marinum_M --MVDELGSNSEVGTLKVVILHRPGTELRRLTPRNTDQLLFDGLPWVSRA
MUL_4664|M.ulcerans_Agy99 --MVDELGSNSEVGTLKVVILHRPGTELRRLTPRSSDQLLFDGLPWVSRA
MAV_1125|M.avium_104 -MGVVELGTNSEVGALRVVILHRPGGELLRLNPRNVDQLLFDGLPWVARA
Mflv_1923|M.gilvum_PYR-GCK MTDAP-LGCNSEVGTLRAVILHRPGAELQRLTPRNNDTLLFDGLPWVARA
Mvan_4810|M.vanbaalenii_PYR-1 MTDAT-LGCNSEVGRLRVVILHRPGPELQRLTPRNNDTLLFDGLPWVARA
MAB_1121|M.abscessus_ATCC_1997 MTDVPVLGANSEVGRLRAVVLHRPGDELKRLTPRNNDQLLFDGLPWVARA
MSMEG_5448|M.smegmatis_MC2_155 MTDVV-LGVDSEVGTLRVAILHRPGPELKRLTPRNNDALLFDGLPWVSKA
TH_0930|M.thermoresistible__bu ------LGLDSEVGRLRVIIVHRPGAELMRLTPQNSDALLFDGLPWVSKA
** :**** *:. ::**** ** **.*:. * *********::*
Rv1001|M.tuberculosis_H37Rv QDEHDEFAELLASRGAEVLLLSDLLTEALHHSGAARMQGIAAAVDAPRLG
Mb1028|M.bovis_AF2122/97 QDEHDEFAELLASRGAEVLLLSDLLTEALHHSGAARMQGIAAAVDAPRLG
MMAR_4492|M.marinum_M QEEHDQFAELLRSRGVEVLLLSELLTEALH-SGAARMQGVAAAVDSRRLG
MUL_4664|M.ulcerans_Agy99 QEEHDQFAELLRSRGVEVLLLSELLTEALH-SGAARMQGVAAAVDSRRLG
MAV_1125|M.avium_104 QEEHDQFAELLRSRGVEVLLLSDLLTEALTHSGAARMQGVAAAVDARRLG
Mflv_1923|M.gilvum_PYR-GCK QQEHDAFADLLRSRGVEVLLLGDLLTEALDKSGAARMQGISAAVDARRLG
Mvan_4810|M.vanbaalenii_PYR-1 QQEHDAFAELLRSRGVEVLLLGVLLTEALSNSGAARMHGISAAVDSRRLG
MAB_1121|M.abscessus_ATCC_1997 QEEHDAFAEVLRSRGVEVLLLSDLLREALA-SGAARVQGIAAAVNARRLG
MSMEG_5448|M.smegmatis_MC2_155 QEEHDQFAEMLRSRGVEVLLVADLLTEALAHSGAARMHGISAAVDARRLG
TH_0930|M.thermoresistible__bu QEEHDEFTALLRSRGVEVLLLADLLTEALGHSGAARMHGIGAAVDPRRLG
*:*** *: :* ***.****:. ** *** *****::*:.***:. ***
Rv1001|M.tuberculosis_H37Rv LPLAQELSAYLRSLDPGRLAHVLTAGMTFNELPSDTR-TDVSLVLRMHHG
Mb1028|M.bovis_AF2122/97 LPLAQELSAYLRSLDPGRLAHVLTAGMTFNELPSDTR-TDVSLVLRMHHG
MMAR_4492|M.marinum_M IPLAQELSAYLRGLDPVRLSHVLTAGMTFNELPADAR-TDVSLVVRMHHD
MUL_4664|M.ulcerans_Agy99 IPLAQELSAYLRGLDPVRLSHVLTAGMTFNELPADAR-TDVSLVVRMHHD
MAV_1125|M.avium_104 VPLAQELSAYLRGLEPARLAQVLMAGMTFNELPADTR-TDVSLVLRMHHG
Mflv_1923|M.gilvum_PYR-GCK APLAQELSAYLRTLEAAPLARILMAGMTFDELPFGE--NELSLVRRMHHG
Mvan_4810|M.vanbaalenii_PYR-1 VPLAQELSAYLRTLDAAALARILMAGMTFDELPFGE--NELSLVRRMHHG
MAB_1121|M.abscessus_ATCC_1997 YALAQDLSAYLRSLDAADLAYVLMCGMTFDELPLGGGVAQPSLVRTMHHG
MSMEG_5448|M.smegmatis_MC2_155 VPLAQELSAHLRTLEPAALAHVLIAGMTFNELPFSG--KELSLVRRMHHG
TH_0930|M.thermoresistible__bu IPLSQELSSYLRTLEPAELAHVLIAGMTFDELPFGPG--ELSLVRRMHHG
.*:*:**::** *:. *: :* .****:*** . : *** ***.
Rv1001|M.tuberculosis_H37Rv GDFVIEPLPNLVFTRDSSIWIGPRVVIPSLALRARVREASLTDLIYAHHP
Mb1028|M.bovis_AF2122/97 GDFVIEPLPNLVFTRDSSIWIGPRVVIPSLALRARVREASLTDLIYAHHP
MMAR_4492|M.marinum_M ADFVIEPLPNLLFTRDSSIWIGPRFVIPSLAMRARVREASLTDIIYAHHP
MUL_4664|M.ulcerans_Agy99 ADFVIEPLPNLLFTRDSSIWIGPRFVIPSLAMRARVREASLTDIIYAHHP
MAV_1125|M.avium_104 GDFVIDPLPNLVFTRDSSIWIGPRVVIPTLALRARVREASLTDLIYAHHP
Mflv_1923|M.gilvum_PYR-GCK GDFVIDPLPNLLFTRDSSFWIGPRVAITSLSMHARVRETSLTDLIYAHHP
Mvan_4810|M.vanbaalenii_PYR-1 ADFVIDPLPNLLFTRDSSFWIGPRVAITSLSMHARVRETSLTDLIYAHHP
MAB_1121|M.abscessus_ATCC_1997 SDFVIEPLPNLLFTRDSSFWIGRKFAITTLALPARVRETSLTDLIYAHHP
MSMEG_5448|M.smegmatis_MC2_155 GDFVIDPLPNLLFTRDSSFWIGPRVAITSLALPARHRETSLTDVIYAHHP
TH_0930|M.thermoresistible__bu GDFVIEPLPNLVFTRDSSFWIGPRVAITSLALPARSRETSLTDVIYAHHP
.****:*****:******:*** :..*.:*:: ** **:****:******
Rv1001|M.tuberculosis_H37Rv RFTGVRRAYESRTAPVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
Mb1028|M.bovis_AF2122/97 RFTGVRRAYESRTAPVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
MMAR_4492|M.marinum_M RFTGIRRAYESRTAPVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
MUL_4664|M.ulcerans_Agy99 RFTGIRRAYESRTAPVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
MAV_1125|M.avium_104 RFTGVRRAYESRTAPVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
Mflv_1923|M.gilvum_PYR-GCK RFLRVRRAYESRSAPIEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
Mvan_4810|M.vanbaalenii_PYR-1 RFLGVRRAYESRSAPIEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
MAB_1121|M.abscessus_ATCC_1997 RFLGVRRAYESHSAPVEGGDVLLLGPGVVAVGVGERTTPAGAEALARSLF
MSMEG_5448|M.smegmatis_MC2_155 RFLGVRRAYESRSAPVEGGDVLLLAPGVVAVGVGERTTPAGAEALARSLF
TH_0930|M.thermoresistible__bu RFLGVRRAYESRSAPVEGGDVLLXX--------HMWADGAEWQCGGCTWF
** :******::**:******* : * :. . : *
Rv1001|M.tuberculosis_H37Rv DDDLAHTVLAVPIAQQRAQMHLDTVCTMVDTDTMVMYANVVDTLEAFTIQ
Mb1028|M.bovis_AF2122/97 DDDLAHTVLAVPIAQQRAQMHLDTVCTMVDTDTMVMYANVVDTLEAFTIQ
MMAR_4492|M.marinum_M DDDLAHTVLAVPIAQRRAQMHLDTVCTMVDVDKVVMYANVVDELTAFTIE
MUL_4664|M.ulcerans_Agy99 DDDLAHTVLAVPIAQRRAQMHLDTVCTMVDVDKVVMYANVVDELTAFTIE
MAV_1125|M.avium_104 DDDLAHTVLAVPIAQRRAQMHLDTVCTMVDTDTVVMYANVVDALSAFTIQ
Mflv_1923|M.gilvum_PYR-GCK DDDLAHTVLAVPIAQERAQMHLDTVCTMVDTDAVVMYPNIQDSLTAFPIR
Mvan_4810|M.vanbaalenii_PYR-1 DDDLAHTVLAVPIAQERAQMHLDTVCTMVDTDAVVMYPNIQDSLTAFTIR
MAB_1121|M.abscessus_ATCC_1997 DDGLAHTVLAVPIAQERASMHLDTVCTMVDTDAVVMYPAIQDSLSAFTIR
MSMEG_5448|M.smegmatis_MC2_155 DDALAHTVLAVPIAQERAQMHLDTVCTMVDVDAVVMYPNIVDSLSAFTIR
TH_0930|M.thermoresistible__bu TSQFTRLEFLDNYDAR------FVVVTNGPIDEALAYREKVGNRMAWYSS
. ::: : . .* * * : * . *:
Rv1001|M.tuberculosis_H37Rv RTPDGVTIGDAAPFAEAAAKAMGIDKLRVIHTGMDPVVAEREQWDDGNNT
Mb1028|M.bovis_AF2122/97 RTPDGVTIGDAAPFAEAAAKAMGIDKLRVIHTGMDPVVAEREQWDDGNNT
MMAR_4492|M.marinum_M RQPDGVTISDAAPFVEAAARAMGIEKLQVIGTGIDPVVAEREQWDDGNNT
MUL_4664|M.ulcerans_Agy99 RQPDGVTISDAAPFVEAAVRAMGIEKLQVIGTGIDPVVAEREQWDDGNNT
MAV_1125|M.avium_104 RTPSGVEIS-EAPFLEAAATAMGIDKLRVIDTGLDPVIAEREQWDDGNNT
Mflv_1923|M.gilvum_PYR-GCK RKSGGVTIDRAAPFVDAAADAMGIGKLRVIDTGLDPVTAEREQWDDGNNT
Mvan_4810|M.vanbaalenii_PYR-1 RESGGVKIDRAAPFVDAAADAMGIAKLRVIDTGLDPVTAEREQWDDGNNT
MAB_1121|M.abscessus_ATCC_1997 RTDSGVSISGAEPFVEAAADAMGIGKLRVIDTGLDPVTAEREQWDDGNNT
MSMEG_5448|M.smegmatis_MC2_155 HDAGGVHISDAQRFVDAAAEAMGIDKLRVIDTGLDPVTAEREQWDDGNNT
TH_0930|M.thermoresistible__bu ADS---PFGADVDAPPGGGFAVNVFLRATAADGTDAVFRTWHTDGRGTEQ
:. .. *:.: . * *.* . . *.:
Rv1001|M.tuberculosis_H37Rv LALAPGVVVAYERNVQTNARLQDAGIEVLTIAGSELGTGRGGPRCMSCPA
Mb1028|M.bovis_AF2122/97 LALAPGVVVAYERNVQTNARLQDAGIEVLTIAGSELGTGRGGPRCMSCPA
MMAR_4492|M.marinum_M LALAPGVVVAYERNAQTNARLEAAGIEVLTIGGSELGTGRGGPRCMSCPV
MUL_4664|M.ulcerans_Agy99 LALAPGVVVAYERNAQTNARLEAAGIEVLTIGGSELGTGRGGPRCMSCPV
MAV_1125|M.avium_104 LALSPGVVVAYERNAQTNMRLQEAGIEVLTIAGSELGTGRGGPRCMSCPV
Mflv_1923|M.gilvum_PYR-GCK LALAPGVVVAYERNTETNARLADSGIEVLPIAASELGTGRGGPRCMSCPA
Mvan_4810|M.vanbaalenii_PYR-1 LAVAPGVVVAYERNTETNARLADSGIEVLPISASELGTGRGGPRCMSCPA
MAB_1121|M.abscessus_ATCC_1997 LALAPGVVVAYERNVETNARLEDSGIEVLAISASELGTGRGGPRCMSCPV
MSMEG_5448|M.smegmatis_MC2_155 LALAPGVVVAYERNSVTNARLQDSGIEVLAIPASELGTGRGGPRCMSCPA
TH_0930|M.thermoresistible__bu LSHVFALLDILPWGRQEEWQDSPDGWPRWPTYSG--WPDSPDIARWYGPA
*: .:: . : : * . .. .. . *.
Rv1001|M.tuberculosis_H37Rv ARDPL-
Mb1028|M.bovis_AF2122/97 ARDPL-
MMAR_4492|M.marinum_M ARDPLP
MUL_4664|M.ulcerans_Agy99 ARDPLP
MAV_1125|M.avium_104 ARDPL-
Mflv_1923|M.gilvum_PYR-GCK GRDPL-
Mvan_4810|M.vanbaalenii_PYR-1 GRDPL-
MAB_1121|M.abscessus_ATCC_1997 SRDLL-
MSMEG_5448|M.smegmatis_MC2_155 ARDPL-
TH_0930|M.thermoresistible__bu PR----
*