For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKVAAQCSKLGYTVAPMEQRAELVVGRALVVVVDDRTAHGDEDHSGPLVTELLTEAGFVVDGVVAVSADE VEIRNALNTAVIGGVDLVVSVGGTGVTPRDVTPEATRDILDREILGIAEAIRASGLSAGIVDAGLSRGLA GVSGSTLVVNLAGSRYAVRDGMATLNPLAAQIIGQLSSLEI
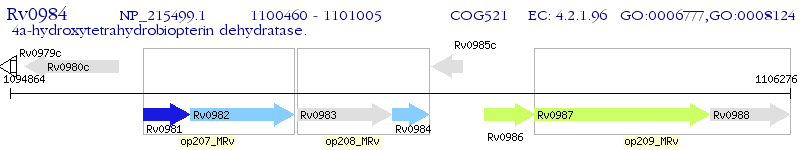
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0984 | moaB2 | - | 100% (181) | POSSIBLE PTERIN-4-ALPHA-CARBINOLAMINE DEHYDRATASE MOAB2 (PHS) (4-ALPHA-HYDROXY-TETRAHYDROPTERIN DEHYDRATASE) (PTERIN-4-A-CARBINOLAMINE DEHYDRATASE) (PHENYLALANINE HYDROXYLASE-STIMULATING PROTEIN) (PHS) (PTERIN CARBINOLAMINE DEHYDRATASE) (PCD) |
| M. tuberculosis H37Rv | Rv0865 | mog | 6e-23 | 41.72% (151) | molybdopterin biosynthesis Mog protein |
| M. tuberculosis H37Rv | Rv1901 | cinA | 5e-05 | 29.03% (93) | competence damage-inducible protein A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1010 | moaB2 | 1e-95 | 100.00% (181) | pterin-4-alpha-carbinolamine dehydratase MoaB2 |
| M. gilvum PYR-GCK | Mflv_1890 | - | 2e-71 | 83.54% (164) | molybdenum cofactor synthesis domain-containing protein |
| M. leprae Br4923 | MLBr_00177 | moaB | 9e-88 | 92.74% (179) | putative molybdenum cofactor biosynthesis protein |
| M. abscessus ATCC 19977 | MAB_1079 | - | 6e-67 | 76.33% (169) | putative molybdopterin biosynthesis protein |
| M. marinum M | MMAR_4526 | moaB2 | 6e-88 | 94.35% (177) | pterin-4-alpha-carbinolamine dehydratase MoaB2 |
| M. avium 104 | MAV_1097 | - | 3e-83 | 96.36% (165) | putative molybdenum cofactor synthesis protein |
| M. smegmatis MC2 155 | MSMEG_5485 | - | 4e-79 | 85.47% (179) | molybdopterin biosynthesis protein |
| M. thermoresistible (build 8) | TH_1731 | moaB2 | 3e-72 | 88.05% (159) | POSSIBLE PTERIN-4-ALPHA-CARBINOLAMINE DEHYDRATASE MOAB2 |
| M. ulcerans Agy99 | MUL_4698 | moaB2 | 2e-88 | 94.35% (177) | pterin-4-alpha-carbinolamine dehydratase MoaB2 |
| M. vanbaalenii PYR-1 | Mvan_4841 | - | 2e-75 | 85.96% (171) | molybdenum cofactor synthesis domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1890|M.gilvum_PYR-GCK ----------------MEQ-PHTLVGRALVVIVDDRTAHGDEEDRTGPLV
Mvan_4841|M.vanbaalenii_PYR-1 --MTAPR----YTVELMEQ-PQALVGRALVVVVDDRTAHGDEEDHSGPLV
MSMEG_5485|M.smegmatis_MC2_155 --MAAPLSPSTYTVADMEQ-PGELVGRALVIVVDDRTAHG-EEDHSGPLV
Rv0984|M.tuberculosis_H37Rv MKVAAQCSKLGYTVAPMEQRAELVVGRALVVVVDDRTAHGDE-DHSGPLV
Mb1010|M.bovis_AF2122/97 MKVAAQCSKLGYTVAPMEQRAELVVGRALVVVVDDRTAHGDE-DHSGPLV
MMAR_4526|M.marinum_M MNADAQLSDLGYSVAPMEQGAELVVGRALVVVVDDRTAHGDE-DHSGPLV
MUL_4698|M.ulcerans_Agy99 MNADAQLSDLGYSVAPMEQGAELVVGRALVVVVDDRTAHGDE-DHSGPLV
MAV_1097|M.avium_104 ----------------METGAELVVGRALVVVVDDRTAHGDE-DHSGPLV
MLBr_00177|M.leprae_Br4923 MLVDKQLSELGYTVAPMEKGVELVVGRALIVVVDDRTAHGDE-DHSGPLV
TH_1731|M.thermoresistible__bu ---------------------VEMVGRALVVVVDDRTAHGDEEDHSGPLV
MAB_1079|M.abscessus_ATCC_1997 ---MTSMSPRTVTVASMAELEAGPVGRSLVVIVNDRTAHGDQ-DTSGPLV
***:*:::*:****** : * :****
Mflv_1890|M.gilvum_PYR-GCK TELLGEAGFVVDGVVVVSSDEVEIRNALNTAVIGGVDLVVSVGGTGVAPR
Mvan_4841|M.vanbaalenii_PYR-1 TELLGEAGFVVDGVVVVSSDEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
MSMEG_5485|M.smegmatis_MC2_155 TELLGEAGFVVDGVVVVASDEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
Rv0984|M.tuberculosis_H37Rv TELLTEAGFVVDGVVAVSADEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
Mb1010|M.bovis_AF2122/97 TELLTEAGFVVDGVVAVSADEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
MMAR_4526|M.marinum_M TELLTEAGFVVDGVVAVEADEVDIRNALNTAVIGGVDLVVSVGGTGVTPR
MUL_4698|M.ulcerans_Agy99 TELLTEAGFVVDGVVAVEADEVDIRNALNTAVIGGVDLVVSVGGTGVTPR
MAV_1097|M.avium_104 TELLTEAGFVVDGVVAVAADEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
MLBr_00177|M.leprae_Br4923 TELLTEAGFVVDGVVAVAADEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
TH_1731|M.thermoresistible__bu TELLNEGGFVVDGVIVVSADEVEIRNALNTAVIGGVDLVVSVGGTGVTPR
MAB_1079|M.abscessus_ATCC_1997 TELLAEAGFVVDGVVVVENDLSEIQNAVNTAVIGGVDLVVTVGGTGVTPR
**** *.*******:.* * :*:**:************:******:**
Mflv_1890|M.gilvum_PYR-GCK DVTPESTVDLLDRELLGISEALRSSGLSAGIIDAGVSRGLAGISGSTLVV
Mvan_4841|M.vanbaalenii_PYR-1 DVTPESTRELLDRELLGIAEALRASGLSAGIIDAGVSRGLAGISGSTLVV
MSMEG_5485|M.smegmatis_MC2_155 DVTPEATRELLDRELLGISEALRASGLAAGIVDAGLSRGLAGISGSTLVV
Rv0984|M.tuberculosis_H37Rv DVTPEATRDILDREILGIAEAIRASGLSAGIVDAGLSRGLAGVSGSTLVV
Mb1010|M.bovis_AF2122/97 DVTPEATRDILDREILGIAEAIRASGLSAGIVDAGLSRGLAGVSGSTLVV
MMAR_4526|M.marinum_M DVTPESTREILDREILGIAEAIRASGLSAGIIDAGLSRGLAGVSGSTLVV
MUL_4698|M.ulcerans_Agy99 DVTPEATREILDREILGIAEAIRASGLSAGIIDAGLSRGLAGVSGSTLVI
MAV_1097|M.avium_104 DVTPEATREILDREILGIAEAIRGSGLSAGIIDAGLSRGLAGVSGSTLVV
MLBr_00177|M.leprae_Br4923 DVAPEATREILDREILGIAEAIRASGLSAGITDAGLSRGLAGVSGSTLVV
TH_1731|M.thermoresistible__bu DVTPEATRELLDRELLGIAEALRASALQAGIADGGVSRGLAGVSGSTLVV
MAB_1079|M.abscessus_ATCC_1997 DVAPEATQPLLDRELLGIAEAIRSSGLAAGVTEAGLSRGVAGISGSTLVV
**:**:* :****:***:**:*.*.* **: :.*:***:**:******:
Mflv_1890|M.gilvum_PYR-GCK NLAGSRAAVRDGMATLGPLAAQIIAQLSSLEI
Mvan_4841|M.vanbaalenii_PYR-1 NLAGSRAAVRDGMATLGPLAGQVIAQLSSLEI
MSMEG_5485|M.smegmatis_MC2_155 NIAGSRAAVRDGMATLGPLAVQIIGQLSSLEI
Rv0984|M.tuberculosis_H37Rv NLAGSRYAVRDGMATLNPLAAQIIGQLSSLEI
Mb1010|M.bovis_AF2122/97 NLAGSRYAVRDGMATLNPLAAQIIGQLSSLEI
MMAR_4526|M.marinum_M NLAGSRYAVRDGMATLNPLAAHIIGQLSSLEI
MUL_4698|M.ulcerans_Agy99 NLAGSRYAVRDGMATLNPLAAHIIGQLSSLEI
MAV_1097|M.avium_104 NLAGSRYAVRDGMATLNPLAAQIIGQLSSLEI
MLBr_00177|M.leprae_Br4923 NLAGSRYAVRDGMATLNPLATQIIGQLSSLEI
TH_1731|M.thermoresistible__bu NLAGSRAAVRDGMATLIPLATHIIGQLSSLEI
MAB_1079|M.abscessus_ATCC_1997 NIAGSRAAVRDGMATLTPMAIQIIEQLSSLEI
*:**** ********* *:* ::* *******