For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKLARVVGLVQEEQPSDMTNHPRYSPPPQQPGTPGYAQGQQQTYSQQFDWRYPPSPPPQPTQYRQPYEA LGGTRPGLIPGVIPTMTPPPGMVRQRPRAGMLAIGAVTIAVVSAGIGGAAASLVGFNRAPAGPSGGPVAA SAAPSIPAANMPPGSVEQVAAKVVPSVVMLETDLGRQSEEGSGIILSAEGLILTNNHVIAAAAKPPLGSP PPKTTVTFSDGRTAPFTVVGADPTSDIAVVRVQGVSGLTPISLGSSSDLRVGQPVLAIGSPLGLEGTVTT GIVSALNRPVSTTGEAGNQNTVLDAIQTDAAINPGNSGGALVNMNAQLVGVNSAIATLGADSADAQSGSI GLGFAIPVDQAKRIADELISTGKASHASLGVQVTNDKDTLGAKIVEVVAGGAAANAGVPKGVVVTKVDDR PINSADALVAAVRSKAPGATVALTFQDPSGGSRTVQVTLGKAEQ
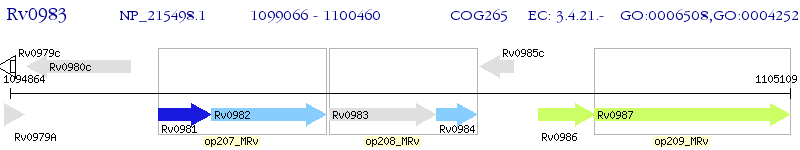
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0983 | pepD | - | 100% (464) | PROBABLE SERINE PROTEASE PEPD (SERINE PROTEINASE) (MTB32B) |
| M. tuberculosis H37Rv | Rv1223 | htrA | 7e-66 | 43.31% (344) | serine protease HtrA |
| M. tuberculosis H37Rv | Rv0125 | pepA | 4e-45 | 36.26% (353) | serine protease PepA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1009 | - | 0.0 | 99.78% (464) | serine protease |
| M. gilvum PYR-GCK | Mflv_1889 | - | 1e-147 | 62.39% (476) | peptidase S1 and S6, chymotrypsin/Hap |
| M. leprae Br4923 | MLBr_00176 | - | 1e-166 | 80.68% (383) | putative secreted serine protease |
| M. abscessus ATCC 19977 | MAB_1078 | - | 1e-130 | 57.52% (452) | putative serine protease |
| M. marinum M | MMAR_4527 | pepD | 0.0 | 76.40% (483) | serine protease PepD |
| M. avium 104 | MAV_1096 | - | 0.0 | 74.54% (491) | protease |
| M. smegmatis MC2 155 | MSMEG_5486 | - | 1e-155 | 67.85% (451) | peptidase S1 and S6, chymotrypsin/Hap |
| M. thermoresistible (build 8) | TH_4473 | - | 1e-135 | 71.79% (358) | PUTATIVE peptidase S1 and S6, chymotrypsin/Hap |
| M. ulcerans Agy99 | MUL_4699 | pepD | 0.0 | 75.78% (483) | serine protease PepD |
| M. vanbaalenii PYR-1 | Mvan_4842 | - | 1e-148 | 63.93% (463) | peptidase S1 and S6, chymotrypsin/Hap |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1889|M.gilvum_PYR-GCK MKMVEAGATGAEPDCCRKGNRTLRPFSVRLGTLSWQRSLFTRQDQFEGEH
Mvan_4842|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4473|M.thermoresistible__bu --------------------------------------------------
MSMEG_5486|M.smegmatis_MC2_155 --------------------------------------------------
Rv0983|M.tuberculosis_H37Rv --------------------------------------------MAKLAR
Mb1009|M.bovis_AF2122/97 --------------------------------------------MAKLAR
MMAR_4527|M.marinum_M --------------------------------------------------
MUL_4699|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00176|M.leprae_Br4923 --------------------------------------------------
MAV_1096|M.avium_104 --------------------------------------------------
MAB_1078|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1889|M.gilvum_PYR-GCK CSDMTNHPRYS--PTPQPGRQSGYTGQVPPGS--AGPAGSGSYEQQGTGS
Mvan_4842|M.vanbaalenii_PYR-1 ---MTNQPSYS--PTPQPGRQSGYPGQVPPGH--AGPR-TGSYEQQGSGG
TH_4473|M.thermoresistible__bu --------------------------------------------------
MSMEG_5486|M.smegmatis_MC2_155 ---MTNHPRYSQQPDPRTGATPGYPGGRPD-------------PYQQTSQ
Rv0983|M.tuberculosis_H37Rv VVGLVQEEQPSDMTNH-PRYSPPPQQPGTP-------GYAQGQQQTYSQQ
Mb1009|M.bovis_AF2122/97 VVGLVQEEQPSDMTNH-PRYSPPPQQPGTP-------GYAQGQQQTYSQQ
MMAR_4527|M.marinum_M ---MTNHPRYSPPPQQ-PGYRTAPNQPVPP-------AYAQGPQQNYNPQ
MUL_4699|M.ulcerans_Agy99 ---MTNHPGYSPPPQQ-PGYRTAPNQPVPP-------AYAQGPQQNYNPQ
MLBr_00176|M.leprae_Br4923 --------------------------------------------------
MAV_1096|M.avium_104 ---MTNDPRYSPPPQQQPGYPPAPNQTAPVPSHAGSSGYAQG-QQPYQQQ
MAB_1078|M.abscessus_ATCC_1997 ---MTNDPHGSWAYRPSGSYSGQHGHHQAP--------NHHAQSSHHYQG
Mflv_1889|M.gilvum_PYR-GCK WDWRYATEQQRQAFRGP-----------------YDSPYDPYRGAPMPGG
Mvan_4842|M.vanbaalenii_PYR-1 WDWRYATEQQRQAFR---------------------APYDPYAGAPMPSG
TH_4473|M.thermoresistible__bu --------------------------------------------------
MSMEG_5486|M.smegmatis_MC2_155 YDWRYAQQVQQ-------------------------APQSPQTHQTQPGF
Rv0983|M.tuberculosis_H37Rv FDWRYPPSPPPQPTQYR-----------------------QPYEALGGTR
Mb1009|M.bovis_AF2122/97 FDWRYPPSPPPQPTQYR-----------------------QPYEALGGTR
MMAR_4527|M.marinum_M FDWRYHQSRPQPRPPYEGVSGTGPGLLPPATGPGPIPPDTGPGPIPGGTG
MUL_4699|M.ulcerans_Agy99 FDWRYHQSRPQPRPPYEGVSGTGPGLLPPATGPGPIPPDTGPGPIPGGTG
MLBr_00176|M.leprae_Br4923 --------------------------------------------------
MAV_1096|M.avium_104 FDWRYR-SQPSQTTQFR--------------SPYEPFGNAGPGRVPGGTG
MAB_1078|M.abscessus_ATCC_1997 QSYGYPSQQPQQ-------------------------------------R
Mflv_1889|M.gilvum_PYR-GCK PGQFPPPGMP--RQPQPQPRKRS-GSALAAGAIALALVSGGIGG--GVAM
Mvan_4842|M.vanbaalenii_PYR-1 PGHHPRPGMP--VTPPRQPRKGSRATALAAGAVAVALVSGGIGG--GVAM
TH_4473|M.thermoresistible__bu ------------------------------------VVSAGIGG--GVAT
MSMEG_5486|M.smegmatis_MC2_155 RPAYDPYRAPSTAQPVAIPAKKRSRAGLAAGALAVAVVSAGIGG--GVAT
Rv0983|M.tuberculosis_H37Rv PGLIPGVIPTMTPPPGMVRQR-PRAGMLAIGAVTIAVVSAGIGG--AAAS
Mb1009|M.bovis_AF2122/97 PGLIPGVIPTMTPPPGMVRQR-PRAGMLAIGAVTIAVVSAGIGG--AAAS
MMAR_4527|M.marinum_M PGPIPGMLPPMAPPPNVIRERRPRAGMLVMGAVVIAVVSAGIGG--AAAS
MUL_4699|M.ulcerans_Agy99 PGPIPGMLPPMAPPPNVIRERRPRAGMLVMGAVVIAVVSAGIGG--AAAS
MLBr_00176|M.leprae_Br4923 ------MIP-----PGRVADRRPRAGMLAVSALAIAVVSAGIGG--VAAT
MAV_1096|M.avium_104 TGPLPGMHPGMPGPPEPQKRS--RAGLLTVGAVAIAVVSAGIGG--AAAT
MAB_1078|M.abscessus_ATCC_1997 PAQQP---------QRQAQPKGRSSGGLVVGAVVLALVSGGLGGTVGAVV
:**.*:** ..
Mflv_1889|M.gilvum_PYR-GCK LAHPDHSLGGIS----ASGAAP-----SMPAASVPGGSVEAVAAKVVPSV
Mvan_4842|M.vanbaalenii_PYR-1 LVHPDHGVPGIS----ASGAAP-----GMPAASVPAGSVEAVAAAVVPSV
TH_4473|M.thermoresistible__bu LAQPERPADYSS----LSDAAS-----AVPAASIPPGTVEQVAAKVVPSV
MSMEG_5486|M.smegmatis_MC2_155 LVHNESPQLGSP----AIGAAP-----TVPAAALPAGSVEQVAAKVVPSV
Rv0983|M.tuberculosis_H37Rv LVGFNRAPAGPSGGPVAASAAP-----SIPAANMPPGSVEQVAAKVVPSV
Mb1009|M.bovis_AF2122/97 LVGFNRAPAGPSGGPVAASAAP-----SIPAANMPPGSVEQVAAKVVPSV
MMAR_4527|M.marinum_M IIELNRSPATSSGREMVASGAP-----SVPAANMPPGSVEQVAAKVLPSV
MUL_4699|M.ulcerans_Agy99 IIELNRSPATSSGREMVASGAP-----SVPAANMPPGSVEQVAAKVLPSV
MLBr_00176|M.leprae_Br4923 VVALGTRVAGGNGAGPVTGPAA-----SVPAANMPSGSVEQVAVKVVPSV
MAV_1096|M.avium_104 AVEMG-THATGSGGGRIAGAAP-----SVPAANMPPGSVEQVAAKVVPSV
MAB_1078|M.abscessus_ATCC_1997 ADRQHKAPVVTSSSTGLTPIAPGRPPAAQPVASLPAGSVEQVAAKVLPSV
*. *.* :* *:** **. *:***
Mflv_1889|M.gilvum_PYR-GCK VKLEVNQGRSTEEGSGVILSSDGLILTNNHVVAAAADNG-----------
Mvan_4842|M.vanbaalenii_PYR-1 VKLEVSQGRASEEGSGVILSTDGLILTNNHVVATAAG-------------
TH_4473|M.thermoresistible__bu VKLETELGRATEEGSGIILSSDGLILTNHHVVSAAVREP-----------
MSMEG_5486|M.smegmatis_MC2_155 VKLETDLGRASEEGSGIILTADGLILTNNHVVAAAKG-------------
Rv0983|M.tuberculosis_H37Rv VMLETDLGRQSEEGSGIILSAEGLILTNNHVIAAAAKP------------
Mb1009|M.bovis_AF2122/97 VMLETDLGRQSEEGSGIILSAEGLILTNNHVIAAAAKP------------
MMAR_4527|M.marinum_M VMLETDVGRQSEEGSGIILSADGLVMTNNHVVAAAAKP------------
MUL_4699|M.ulcerans_Agy99 VMLETDVGRQSEEGSGIILSADGLVMTNNHVVAAAAKP------------
MLBr_00176|M.leprae_Br4923 VMLETDLGRQSEEGSGVILSADGLILTNNHVVAVAAKPG-----------
MAV_1096|M.avium_104 VMLETDLGRQSEEGSGIILSTDGLILTNNHVVAAAAGPPKGPAAPPVVPL
MAB_1078|M.abscessus_ATCC_1997 VKLETIMGRSSEEGSGVILSADGLVLTNAHVVQAAGDPG-----------
* **. ** :*****:**:::**::** **: .*
Mflv_1889|M.gilvum_PYR-GCK ----APGAPPGA-AQTKVTFADGKSSTFEVIGTDPGSDIAVVRAKNVTDL
Mvan_4842|M.vanbaalenii_PYR-1 ----AAGEPGGP-AKTKVTFADGKTAPFTVIGADPSSDIAVVRAQNVSGL
TH_4473|M.thermoresistible__bu ----GRTDAQTPNVRTKVTFADGRTAPFTVVGSDPSSDIAVVRATNVTGL
MSMEG_5486|M.smegmatis_MC2_155 -------DGPGPAAQTKVTFADGTTRPFTVVGTDPSSDIAVVRAQDVSGL
Rv0983|M.tuberculosis_H37Rv ----PLGSP---PPKTTVTFSDGRTAPFTVVGADPTSDIAVVRVQGVSGL
Mb1009|M.bovis_AF2122/97 ----PLGSP---PPKTTVTFSDGRTAPFTVVGADPTSDIAVVRVQGVSGL
MMAR_4527|M.marinum_M ----PAGAP---EPKTTVTLYDGRTASFTVVGADPTSDIAVVRVSGVSGL
MUL_4699|M.ulcerans_Agy99 ----PAGAP---EPKTTVTLYDGRTASFTVVGADPTSDIAVVRVSGVSGL
MLBr_00176|M.leprae_Br4923 --GGPGGGL---SPKTTVTFFDGRTASFTVVGADPTSDIAVVRVQSISGL
MAV_1096|M.avium_104 PPGGPGGGPGAGTPKTTVTFSDGRTAPFTVVGADPTSDIAVIRVQGMSGL
MAB_1078|M.abscessus_ATCC_1997 -------------AKTTATIAGGKTSPFTVVGIDPGTDIAVVRITGASGL
:*..*: .* : .* *:* ** :****:* . :.*
Mflv_1889|M.gilvum_PYR-GCK TPIAVGSSADLRVGQDVVAIGSPLGLEGTVTTGIISALNRPVAASGDARN
Mvan_4842|M.vanbaalenii_PYR-1 TPITVGSSADLRVGQDVVAIGSPLGLEGTVTTGIISALNRPVAAGGDARN
TH_4473|M.thermoresistible__bu TPVTLGSSADLRVGQDVVAIGSPLGLEGTVTTGIVSALNRPVAAGGDARN
MSMEG_5486|M.smegmatis_MC2_155 TPITLGSSANLRVGQDVVAIGSPLGLEGTVTTGIVSALNRPVAAGGDTRN
Rv0983|M.tuberculosis_H37Rv TPISLGSSSDLRVGQPVLAIGSPLGLEGTVTTGIVSALNRPVSTTGEAGN
Mb1009|M.bovis_AF2122/97 TPISLGSSSDLRVGQPVLAIGSPLGLEGTVTTGIVSALNRPVSTTGEAGN
MMAR_4527|M.marinum_M TPIALGSSADLRVGQPVLAIGSPLGLAGTVTTGIVSALNRPVSTTGEAGN
MUL_4699|M.ulcerans_Agy99 TPIALGSSADLRVGQPLLAIGSPLGLAGTVTTGIVSALNRPVSTTGESGN
MLBr_00176|M.leprae_Br4923 TPITMGSSADLRVGQPVVAVGSPLGLAGTVTSGIVSALNRPVSTTGESGN
MAV_1096|M.avium_104 TPISLGSSSDLRVGQPVVAIGSPLGLSGTVTTGIVSALNRPVSTTGESGN
MAB_1078|M.abscessus_ATCC_1997 TPIALGSSANLVVGQSVVAIGSPLGLDGTVTTGIVSSLNRPVFTAGQAGN
**:::***::* *** ::*:****** ****:**:*:***** : *:: *
Mflv_1889|M.gilvum_PYR-GCK QNTVLDAIQTDAAINPGNSGGALVNMNGELVGINSAIATMGADA-GGPQG
Mvan_4842|M.vanbaalenii_PYR-1 QNTVLDAIQTDAAINPGNSGGALVNMNGELVGINSAIATMGADA-GAQQG
TH_4473|M.thermoresistible__bu QNTVLDAIQTDAAINPGNSGGALVNMNGELVGVNSAIATLGGGV-AQTQS
MSMEG_5486|M.smegmatis_MC2_155 QNTVLDAIQTDAAINPGNSGGALVNMNGELVGINSAIATMGGDS-PNAQS
Rv0983|M.tuberculosis_H37Rv QNTVLDAIQTDAAINPGNSGGALVNMNAQLVGVNSAIATLGADS-ADAQS
Mb1009|M.bovis_AF2122/97 QNTVLDAIQTDAAINPGNSGGALVNMNAQLVGVNSAIATLGADS-ADAQS
MMAR_4527|M.marinum_M QNTVLDAIQTDAAINPGNSGGALVNMNAQLVGINSAIATLGADS-GDAQS
MUL_4699|M.ulcerans_Agy99 QNTVLDAIQTDAAINPGNSGGALVNMNAQLVGINSAIATLGADS-GDAQS
MLBr_00176|M.leprae_Br4923 QNTVLDAIQTDAAINPGNSGGALVNMGGQLVGVNSAIATLGADS-GDAQS
MAV_1096|M.avium_104 QNTVLDAIQTDAAINPGNSGGALVNMNGQLVGVNSAIATLGADS-PDAQS
MAB_1078|M.abscessus_ATCC_1997 QDTVLDAIQTDAAINPGNSGGALVNLNGELVGINSAIATMGGDSSGETQG
*:***********************:..:***:******:*.. *.
Mflv_1889|M.gilvum_PYR-GCK GSIGLGFAIPVDQAKRIADEIIQTGSASRASLGVQVGNEAGVDGAKIVEV
Mvan_4842|M.vanbaalenii_PYR-1 GSIGLGFAIPVDQAKRIADEIIQTGSASRASLGVQVGNEAGVDGAKIVEV
TH_4473|M.thermoresistible__bu GSIGLGFAIPVDQAKRIADELIKHGRATHASLGVQVSNEAGTDGAMIVEV
MSMEG_5486|M.smegmatis_MC2_155 GSIGLGFAIPVDQAKRIADELIQNGVASHASLGVQVANDRSSDGAKIVEV
Rv0983|M.tuberculosis_H37Rv GSIGLGFAIPVDQAKRIADELISTGKASHASLGVQVTNDKDTLGAKIVEV
Mb1009|M.bovis_AF2122/97 GSIGLGFAIPVDQAKRIADELISTGKASHASLGVQVTNDKDTPGAKIVEV
MMAR_4527|M.marinum_M GSIGLGFAIPVDQAKRIADELISTGKASHASLGVQVTTDKGVPGAKVVEV
MUL_4699|M.ulcerans_Agy99 GSIGLGFAIPVDQAKRIADELISTGKASHASLGVQVTTDKGVPGAKVVEV
MLBr_00176|M.leprae_Br4923 GSIGLGFAIPVDQAKRIADELISTGKATHASLGVQVATDKGTPGAKVMDV
MAV_1096|M.avium_104 GSIGLGFAIPVDQAKRIADELIATGKASHASLGVQVTNDKGTPGAKVVDV
MAB_1078|M.abscessus_ATCC_1997 GSIGLGFAIPVDQAKRVADQLITNGRATHASLGVRVGNDKEVQGARIVEV
****************:**::* * *::*****:* .: ** :::*
Mflv_1889|M.gilvum_PYR-GCK TAGGAASAAGLPSGVIVTKLDDRAISSADALVAAVRSKAPGDKVTLTYQD
Mvan_4842|M.vanbaalenii_PYR-1 TAGGAASAAGLPSGVIVTKVDDRLINSADALVAAVRSKAPGDKVTLTYLD
TH_4473|M.thermoresistible__bu TEGGAAAAAGLPAGVVVTKLDDRLINSADALVAAVRSKAPGDTVTLTYVE
MSMEG_5486|M.smegmatis_MC2_155 NDGGAAAAAGLPSGVVVTKVDDRVIGSADALVAAVRSKAPGEKVTLTYLD
Rv0983|M.tuberculosis_H37Rv VAGGAAANAGVPKGVVVTKVDDRPINSADALVAAVRSKAPGATVALTFQD
Mb1009|M.bovis_AF2122/97 VAGGAAANAGVPKGVVVTKVDDRPINSADALVAAVRSKAPGATVALTFQD
MMAR_4527|M.marinum_M VQGGAAATAGLPKGVVVTKVDDRAITSADALVAAVRSKAPGDKVSLTFQD
MUL_4699|M.ulcerans_Agy99 VQGGAAATAGLPKGVVVTKVDDRAITSADALVAAVRSKAPGDKVSLTFQD
MLBr_00176|M.leprae_Br4923 VAGGAAANAAVPKGVVLTKVDDRLISSADALVAAVRSKAPGDKVSLTYQD
MAV_1096|M.avium_104 VSGGAAASAGLPKNVVVTKVDDRPINSADALVAAVRSKAPGDKITLTFSD
MAB_1078|M.abscessus_ATCC_1997 VAGGAAEKAGIPNGAVVTKVDNRPVNSADGLVAAVRSKAPGDKVTLTYTD
**** *.:* ..::**:*:* : ***.*********** .::**: :
Mflv_1889|M.gilvum_PYR-GCK ASGK-PRTVDVTLGKAKQ
Mvan_4842|M.vanbaalenii_PYR-1 PAGK-SQSLDVTLGKAGQ
TH_4473|M.thermoresistible__bu GSGK-PRTVDVTLGKMQR
MSMEG_5486|M.smegmatis_MC2_155 QAGK-PQTVDVTLGKAEQ
Rv0983|M.tuberculosis_H37Rv PSGG-SRTVQVTLGKAEQ
Mb1009|M.bovis_AF2122/97 PSGG-SRTVQVTLGKAEQ
MMAR_4527|M.marinum_M PDGS-TRTVQVTLGKAEQ
MUL_4699|M.ulcerans_Agy99 PDGS-TRTVQVTLGKAEQ
MLBr_00176|M.leprae_Br4923 QSGS-SRTVQVTLGKAEQ
MAV_1096|M.avium_104 PAGGGSRTLPVTLGKADQ
MAB_1078|M.abscessus_ATCC_1997 GNGGQEKTADVTLGEVAR
* :: ****: :