For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAATAHGLCEFIDASPSPFHVCATVAGRLLGAGYRELREADRWPDKPGRYFTVRAGSLVAWNAEQSGHTQ VPFRIVGAHTDSPNLRVKQHPDRLVAGWHVVALQPYGGVWLHSWLDRDLGISGRLSVRDGTGVSHRLVLI DDPILRVPQLAIHLAEDRKSLTLDPQRHINAVWGVGERVESFVGYVAQRAGVAAADVLAADLMTHDLTPS ALIGASVNGTASLLSAPRLDNQASCYAGMEALLAVDVDSASSGFVPVLAIFDHEEVGSASGHGAQSDLLS SVLERIVLAAGGTREDFLRRLTTSMLASADMAHATHPNYPDRHEPSHPIEVNAGPVLKVHPNLRYATDGR TAAAFALACQRAGVPMQRYEHRADLPCGSTIGPLAAARTGIPTVDVGAAQLAMHSARELMGAHDVAAYSA ALQAFLSAELSEA
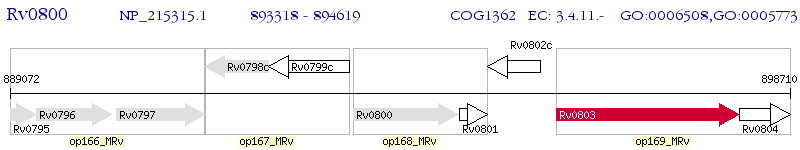
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0800 | pepC | - | 100% (433) | putative aminopeptidase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0823 | pepC | 0.0 | 99.77% (433) | putative aminopeptidase 2 |
| M. gilvum PYR-GCK | Mflv_1628 | - | 1e-180 | 72.83% (427) | putative aminopeptidase 2 |
| M. leprae Br4923 | MLBr_02213 | pepC | 0.0 | 78.29% (433) | putative aminopeptidase 2 |
| M. abscessus ATCC 19977 | MAB_0701 | - | 1e-160 | 66.36% (428) | putative aminopeptidase 2 |
| M. marinum M | MMAR_4891 | pepC | 0.0 | 79.35% (431) | aminopeptidase PepC |
| M. avium 104 | MAV_0742 | - | 0.0 | 83.33% (426) | putative aminopeptidase 2 |
| M. smegmatis MC2 155 | MSMEG_5828 | - | 0.0 | 74.65% (430) | putative aminopeptidase 2 |
| M. thermoresistible (build 8) | TH_0570 | pepC | 0.0 | 75.41% (427) | PROBABLE AMINOPEPTIDASE PEPC |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5121 | - | 1e-180 | 74.00% (427) | putative aminopeptidase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1628|M.gilvum_PYR-GCK -----------------MSASPQGLCEFIDASPSPFHACRTAADRLLAAG
Mvan_5121|M.vanbaalenii_PYR-1 -----------------MPASPQGLCEFIDASPSPFHVCRTAAERLLAAG
MSMEG_5828|M.smegmatis_MC2_155 -----------------MAASPHSLCEFIDASPSPFHVCATAAARLRDAG
TH_0570|M.thermoresistible__bu -----------------MSASAEGLCEFIDASPSPYHVCATVADRLRAAG
Rv0800|M.tuberculosis_H37Rv -----------------MAATAHGLCEFIDASPSPFHVCATVAGRLLGAG
Mb0823|M.bovis_AF2122/97 -----------------MAATAHGLCEFIDASPSPFHVCATVAGRLLGAG
MAV_0742|M.avium_104 MFTMSGGPRNQTK----SSASAQGLCEFIDASPSPFHVCATVAARLIAAG
MLBr_02213|M.leprae_Br4923 MPTLASTTSSPTAMMLGMPASAADLCEFINASPSPFHVCATVAGRLLDAG
MMAR_4891|M.marinum_M MSDAPTTS-----------ATARGLGEFIDASPSPFHLCATVARRLLDAG
MAB_0701|M.abscessus_ATCC_1997 -----------------MTATAVGLCQFIDASPSPFHAVDTVAQRLRHEG
*:. .* :**:*****:* *.* ** *
Mflv_1628|M.gilvum_PYR-GCK FTELAESDPWP-AAGDFFAVRAGSLIAWRS-NTDSAAAPFRVVGAHTDSP
Mvan_5121|M.vanbaalenii_PYR-1 FTELSESDPWP-GAGDHFAVRAGSLIAWRS-PDGSRALPFRIVGAHTDSP
MSMEG_5828|M.smegmatis_MC2_155 YTELAETDAWP-AAGRFFTVRAGSLVAWRT-VED-ASAPFRIVGGHTDSP
TH_0570|M.thermoresistible__bu FVELLERDPWPGAAGDYFTVRAGSLIAWRT-TAD-TGAGFRIVGAHTDSP
Rv0800|M.tuberculosis_H37Rv YRELREADRWPDKPGRYFTVRAGSLVAWNAEQSGHTQVPFRIVGAHTDSP
Mb0823|M.bovis_AF2122/97 YRELREADRWPDKPGRYFTVRAGSLVAWNAEQSGHTQVPFRIVGAHTDSP
MAV_0742|M.avium_104 YTELDEAKHWPSQPGRYFTVRAGSLVAWNSTGADG---PFRIVGAHTDSP
MLBr_02213|M.leprae_Br4923 YAELSEVERWPDHPGRYFIVRAGSLVAWSAGQGVKAHAPFRIVGAHTDSP
MMAR_4891|M.marinum_M YTELSEIDRWPQQPGRYFTVRAGSLVAWDSGGVGTAQAPFRIVGAHSDSP
MAB_0701|M.abscessus_ATCC_1997 FTELFEAQRWPGHSGDYFVVRSGSIVAWRGVDMEGEHAAFRVIGAHTDSP
: ** * . ** .* .* **:**::** **::*.*:***
Mflv_1628|M.gilvum_PYR-GCK NLRVKQHPDRFLSGWQIVALQPYGGAWLNSWLDRDLGVSGRLSVRD---G
Mvan_5121|M.vanbaalenii_PYR-1 NLRVKQHPDRFVAGWQVVALQPYGGAWLNSWLDRDLGVSGRLSVRD---G
MSMEG_5828|M.smegmatis_MC2_155 NLRVKQRPDRMVAGWQVVALQPYGGAWLNSWLDRDLGISGRLTLRDESAD
TH_0570|M.thermoresistible__bu NLRVKEHPDRFVAGWRVVALQPYGGVWLNSWLDRDLGLSGRLSVRS---G
Rv0800|M.tuberculosis_H37Rv NLRVKQHPDRLVAGWHVVALQPYGGVWLHSWLDRDLGISGRLSVRD---G
Mb0823|M.bovis_AF2122/97 NLRVKQHPDRLVAGWHVVALQPYGGVWLHSWLDRDLGISGRLSVRD---G
MAV_0742|M.avium_104 NLRVKQHPDRTVAGWRVVALQPYGGAWLNSWLDRDLGISGRLSVRD---G
MLBr_02213|M.leprae_Br4923 NLRVKQHPDLLVAGWRVVALQPYGGAWLNSWLDRDLGVSGRLSVRSAGKG
MMAR_4891|M.marinum_M NLRVKQHPDRLVAGWQVVALEPYGGAWLNSWLDRDLGVSGRLSVRAPGVR
MAB_0701|M.abscessus_ATCC_1997 NLRVKANADRVQAGWPMVALEPYGGAWVQTWLDRDLGLSGRVLVRD---G
***** ..* :** :***:****.*:::*******:***: :*
Mflv_1628|M.gilvum_PYR-GCK NTVRHLLVRVDDPILRVPQLAIHLS--EDRKGVELNPQRHVNAVWGVGGG
Mvan_5121|M.vanbaalenii_PYR-1 NSIRHTLVRVDDPILRVPQLAIHLS--EDRKGVELNPQRHVNAVWGTGTG
MSMEG_5828|M.smegmatis_MC2_155 DGIAHHLVRIDDPILRVPQLAIHLS--DDRKGVSPDPQRHLNGVWGLGER
TH_0570|M.thermoresistible__bu SGLEHRLIRIDEPVLRVPQLAIHLA--EDRKSVTLNPQQHVNAVWGVADR
Rv0800|M.tuberculosis_H37Rv TGVSHRLVLIDDPILRVPQLAIHLA--EDRKSLTLDPQRHINAVWGVGER
Mb0823|M.bovis_AF2122/97 TGVSHRLVRIDDPILRVPQLAIHLA--EDRKSLTLDPQRHINAVWGVGER
MAV_0742|M.avium_104 SGLSHRLVRIDEPILRVPQLAIHLA--EDRKSVTLDPQRHVNAVWGVGES
MLBr_02213|M.leprae_Br4923 SEITDRLVRIDDPILRVPQLAIHLA--EDRKSLTLDPQRHVNAVWGVGDK
MMAR_4891|M.marinum_M GGISHRLVRIDEPILRVPQLAIHLA--EDRKSLTLDPQRHVNAVWGVGAQ
MAB_0701|M.abscessus_ATCC_1997 DAVRSTLIRIDQPILRVPNLAIHMVSAEERKKWVIDPQWHVNSVWSG---
: *: :*:*:****:****: ::** :** *:*.**.
Mflv_1628|M.gilvum_PYR-GCK ARSFLGYIAGRAGVTEDDVLGFDLMTHDLAPSRLVGAD----GELVSAPR
Mvan_5121|M.vanbaalenii_PYR-1 ARSFLDFVAGRAGVVGDDVLGFDLMTHDLTPSRLAGVD----DNLVSAPR
MSMEG_5828|M.smegmatis_MC2_155 PGVFIEFVADRAGVDAADVLGFDLMTHDLAPSAVTGAA----GEFVSAPR
TH_0570|M.thermoresistible__bu DRAFLDHVADHAGVAAADVLGFDLMTHDLAPSQLIGSD----RDLISAPR
Rv0800|M.tuberculosis_H37Rv VESFVGYVAQRAGVAAADVLAADLMTHDLTPSALIGASVNGTASLLSAPR
Mb0823|M.bovis_AF2122/97 VESFVGYVAQRAGVAAADVLAADLMTHDLTPSALIGASVNGTASLLSAPR
MAV_0742|M.avium_104 ARSFVDYVAARAAVAPDDVLAADLMTHDLTPSTLIGASENGTADLLSAPR
MLBr_02213|M.leprae_Br4923 AGSLLEYVAERTGVAVADVLAVDLMTHDLVPSMVIGAD----ANLLSAPR
MMAR_4891|M.marinum_M RGSFLGYVAQRAGLAEGDVLAADLMTHDLTPAAVIGAN----ADLLSAPR
MAB_0701|M.abscessus_ATCC_1997 ELGFEEYIAEQLGVSPADVLGRDLMTHDLTGAALVGPD----ESLLSAPR
: .:* : .: ***. *******. : : * .::****
Mflv_1628|M.gilvum_PYR-GCK LDNQATCYAGLEAFLAAEP----DGVVPVLVLFDHEEVGSQSDHGAQSDL
Mvan_5121|M.vanbaalenii_PYR-1 LDNQATCYAGLEAFLSARAG---ADAVPVLVLFDHEEVGSQSDHGAQSEL
MSMEG_5828|M.smegmatis_MC2_155 LDNQATCYAGLEAFLAAEE----SGYLPVLALFDHEEVGSQSDHGAQSEL
TH_0570|M.thermoresistible__bu LDNQASCYAGLEALLSVEP----GDHLPVLALFDHEEVGSTSDAGAQSEL
Rv0800|M.tuberculosis_H37Rv LDNQASCYAGMEALLAVDVDSASSGFVPVLAIFDHEEVGSASGHGAQSDL
Mb0823|M.bovis_AF2122/97 LDNQASCYAGMEALLAVDVDSASSGFVPVLAIFDHEEVGSASGHGAQSDL
MAV_0742|M.avium_104 LDNQASCYAGLEALLATEP----SGPLPVLVLFDHEEVGSSSDHGAQSDL
MLBr_02213|M.leprae_Br4923 LDNQVSCYAGMEALLASVP----HDCLPVLALFDHEEVGSTSDRGARSNL
MMAR_4891|M.marinum_M LDNQASCYAGLEALLAAES----DQVLPVLAIFDHEEVGSTSDHGAQSNL
MAB_0701|M.abscessus_ATCC_1997 LDNQASCYAGLEAFLAAES-----SSTTVLALFDHEEVGSVSDHGAHSDL
****.:****:**:*: .**.:******** *. **:*:*
Mflv_1628|M.gilvum_PYR-GCK LLTTLERVTLAAGGSREDFLRRLPDSVVASGDMAHATHPNYPERHEPGHL
Mvan_5121|M.vanbaalenii_PYR-1 LLTVLERITLAAGGSREDFLRRLPASMVASGDMAHATHPNYPDRHEPGHL
MSMEG_5828|M.smegmatis_MC2_155 LPTVLERIALAAGQSREDFLRRVAGSMVASGDMAHATHPNYPERHEPGHL
TH_0570|M.thermoresistible__bu LLTVLERITLAAGGTREDFLRRLPGSMVASGDMAHATHPNYPDRHEPGHL
Rv0800|M.tuberculosis_H37Rv LSSVLERIVLAAGGTREDFLRRLTTSMLASADMAHATHPNYPDRHEPSHP
Mb0823|M.bovis_AF2122/97 LSSVLERIVLAAGGTREDFLRRLTTSMLASADMAHATHPNYPDRHEPSHP
MAV_0742|M.avium_104 LSTVLERIVLAAGGTREDYLSRLPASLLASADMAHATHPNYPERHEPGHP
MLBr_02213|M.leprae_Br4923 LSTVLERIVLAAGGGRDDYLRRLPASLLVSADMAHATHPNYPECHEPSHL
MMAR_4891|M.marinum_M LNTVLERIVLAGGGVREDLLRLLPASLLASADMAHATHPNYPERHEPSHQ
MAB_0701|M.abscessus_ATCC_1997 LSTVLERIVLSSNGTREDFLRRAAASLMVSADMAHATHPNYPDRHEPGHQ
* :.***:.*:.. *:* * . *::.*.***********: ***.*
Mflv_1628|M.gilvum_PYR-GCK IEVNAGPVLKVQPNLRYATDGRTAATFALACAQAGVNLQRYEHRADLPCG
Mvan_5121|M.vanbaalenii_PYR-1 IEVNAGPVLKVQPNLRYATDGRTAAAFALACAQAGVNLQRYEHRADLPCG
MSMEG_5828|M.smegmatis_MC2_155 IEVNAGPVLKVQPNLRYATDGRTAAAFALACDQAGVPLQRYEHRADLPCG
TH_0570|M.thermoresistible__bu IEVNGGPVLKVQPNLRYATDGRTAAAFALACQQAGVPLQRYEHRADLPCG
Rv0800|M.tuberculosis_H37Rv IEVNAGPVLKVHPNLRYATDGRTAAAFALACQRAGVPMQRYEHRADLPCG
Mb0823|M.bovis_AF2122/97 IEVNAGPVLKVHPNLRYATDGRTAAAFALACQRAGVPMQRYEHRADLPCG
MAV_0742|M.avium_104 IAVNAGPVLKVHPNLRYATDSRTAAAFALACRQAGVTLQRYEHRADLPCG
MLBr_02213|M.leprae_Br4923 IEVNAGPVLKVHPNLRYATDGRTAAAFEVACQQAGVRLQRYEHRADRPCG
MMAR_4891|M.marinum_M IAVNAGPVLKVHPNLRYATEGRTAAAFALACQQAGVNLQRYEHRADLPCG
MAB_0701|M.abscessus_ATCC_1997 IAVNAGPVIKIHPNVRYATDGRGAAAFALACERAGVPVQRYEHRADMQCG
* **.***:*::**:****:.* **:* :** :*** :******** **
Mflv_1628|M.gilvum_PYR-GCK STIGPMTAANTGIPTVDVGAAQLAMHSAREVMGAGDVADYSAALQAFLSP
Mvan_5121|M.vanbaalenii_PYR-1 STIGPMTAANTGIPTVDVGAAQLAMHSAREVMGAADVADYSAALAAFLSP
MSMEG_5828|M.smegmatis_MC2_155 STIGPMTAARTGIPTVDVGAAQLAMHSAREFMGAHDVAAYSAALQAFLSP
TH_0570|M.thermoresistible__bu STIGPMTSARTGIPTVDVGAPQLAMHSARELMGAADVTSYSAALRAFLSP
Rv0800|M.tuberculosis_H37Rv STIGPLAAARTGIPTVDVGAAQLAMHSARELMGAHDVAAYSAALQAFLSA
Mb0823|M.bovis_AF2122/97 STIGPLAAARTGIPTVDVGAAQLAMHSARELMGAHDVAAYSAALQAFLSA
MAV_0742|M.avium_104 STIGPLAAARTGIPTVDVGAAQLAMHSARELMGAHDVAAYSAAMHAFLSP
MLBr_02213|M.leprae_Br4923 STIGPLASARTGIPTVDVGAAQLAMHSARELMGAHDVAVYSAALQAFFSA
MMAR_4891|M.marinum_M STIGPMTSARTGIPTVDVGAAQLAMHSARELMGAADVAAYSAALRAFYAP
MAB_0701|M.abscessus_ATCC_1997 STIGPFAASHTGITTVDVGAATLAMHSVRELMGAHDVPMYADALQAFLEL
*****::::.***.******. *****.**.*** **. *: *: **
Mflv_1628|M.gilvum_PYR-GCK V----
Mvan_5121|M.vanbaalenii_PYR-1 G----
MSMEG_5828|M.smegmatis_MC2_155 A----
TH_0570|M.thermoresistible__bu A----
Rv0800|M.tuberculosis_H37Rv ELSEA
Mb0823|M.bovis_AF2122/97 ELSEA
MAV_0742|M.avium_104 G----
MLBr_02213|M.leprae_Br4923 DLF--
MMAR_4891|M.marinum_M E----
MAB_0701|M.abscessus_ATCC_1997 PATS-