For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTLKLHDLRPARGSKIARTRVGRGDGSKGKTAGRGTKGTRARKQVPVTFEGGQMPIHMRLPKLKGFRNRF RTEYEIVNVGDINRLFPQGGAVGVDDLVAKGAVRKNALVKVLGDGKLTAKVDVSAHKFSGSARAKITAAG GSATEL
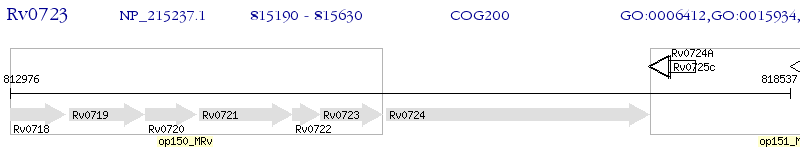
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0723 | rplO | - | 100% (146) | 50S ribosomal protein L15 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0744 | rplO | 4e-79 | 99.32% (146) | 50S ribosomal protein L15 |
| M. gilvum PYR-GCK | Mflv_5028 | rplO | 2e-66 | 81.94% (144) | 50S ribosomal protein L15 |
| M. leprae Br4923 | MLBr_01840 | rplO | 9e-68 | 82.19% (146) | 50S ribosomal protein L15 |
| M. abscessus ATCC 19977 | MAB_3793c | rplO | 9e-62 | 78.08% (146) | 50S ribosomal protein L15 |
| M. marinum M | MMAR_1054 | rplO | 3e-71 | 88.36% (146) | 50S ribosomal protein L15, RplO |
| M. avium 104 | MAV_4446 | rplO | 1e-72 | 89.04% (146) | 50S ribosomal protein L15 |
| M. smegmatis MC2 155 | MSMEG_1474 | rplO | 7e-67 | 83.33% (144) | 50S ribosomal protein L15 |
| M. thermoresistible (build 8) | TH_0304 | rplO | 6e-70 | 85.71% (147) | PROBABLE 50S RIBOSOMAL PROTEIN L15 RPLO |
| M. ulcerans Agy99 | MUL_0813 | rplO | 3e-71 | 87.67% (146) | 50S ribosomal protein L15 |
| M. vanbaalenii PYR-1 | Mvan_1341 | rplO | 2e-66 | 84.03% (144) | 50S ribosomal protein L15 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5028|M.gilvum_PYR-GCK MSDPIKLHDLRPAPGEKTKKTRVGRGEGS-KGKTAGRGTKGTKARKNVPV
Mvan_1341|M.vanbaalenii_PYR-1 -MDPIKLHDLRPAPGEKKAKTRVGRGEGS-KGKTAGRGTKGTKARKNVPV
MSMEG_1474|M.smegmatis_MC2_155 -MSVIKLHDLKPAPGEKKAKTRVGRGEGS-KGKTAGRGTKGTKARKNVPV
TH_0304|M.thermoresistible__bu --MTIKLHDLRPAAGSKTSKTRVGRGDGSGRGKTAGRGTKGTKARKNVPV
Rv0723|M.tuberculosis_H37Rv --MTLKLHDLRPARGSKIARTRVGRGDGS-KGKTAGRGTKGTRARKQVPV
Mb0744|M.bovis_AF2122/97 --MTLKLHDLRPARGSKTARTRVGRGDGS-KGKTAGRGTKGTRARKQVPV
MAV_4446|M.avium_104 --MTIKLHDLKPARGSKTPRTRVGRGEGS-KGKTAGRGTKGTKARKNVPV
MMAR_1054|M.marinum_M --MTIKLHDLRPAPGSKTPRTRVGRGEGS-KGKTAGRGTKGTKARKQVPT
MUL_0813|M.ulcerans_Agy99 --MTIKLHDLRPAPGSKTPRTRVGRGEGS-KGKTAGRGTKGTKARKQVPT
MLBr_01840|M.leprae_Br4923 --MTIKLHDLQPARGSKTTRTRVGRGEAS-KGKTAGRGTKGTKARKQVPV
MAB_3793c|M.abscessus_ATCC_199 --MTIKLHHLRPAPGSKTERTRVGRGEGS-KGKTAGRGTKGTKARKNVPV
:***.*:** *.* :******:.* :***********:***:**.
Mflv_5028|M.gilvum_PYR-GCK MFEGGQMPIHMRLPKLKGFRNRFRTEYGVVNVGDLNKAFPEGGTVGVDEL
Mvan_1341|M.vanbaalenii_PYR-1 TFEGGQMPIHMRLPKLKGFRNRFRTEYAPVNVGDIAKAFPEGGTVGVDEL
MSMEG_1474|M.smegmatis_MC2_155 MFEGGQMPIHMRLPKLKGFKNRFRTEYQVVNVGDINKAFPQGGTVGVDEL
TH_0304|M.thermoresistible__bu TFEGGQMPIHMRLPKLRGFRNRFRTEYAVVNVGDINRLFPEGGQVGIDEL
Rv0723|M.tuberculosis_H37Rv TFEGGQMPIHMRLPKLKGFRNRFRTEYEIVNVGDINRLFPQGGAVGVDDL
Mb0744|M.bovis_AF2122/97 TFEGGQMPIHMRLPKLKGFRNRFRTEYEIVNVGDINRLFPQGGAVGVDDL
MAV_4446|M.avium_104 TFEGGQMPIHMRLPKLKGFRNRFRTEYEIVNVGDINRLFPQGGSVGVDEL
MMAR_1054|M.marinum_M TFEGGQMPIHMRLPKLKGFRNRFRTEYEIVNVGDIARLFPEGGTVGVDEL
MUL_0813|M.ulcerans_Agy99 TFEGGQMPIHMRLPKLKGFRNRFRTEYEIVNVGDIARLFPEGGTVGVDEL
MLBr_01840|M.leprae_Br4923 TFEGGQMPIHMRLPKLKGFRNRLRTEYAVVNVGDISRLFPEGGTISVNDL
MAB_3793c|M.abscessus_ATCC_199 TFEGGQMPIHMRLPKLKGFKNRFRTEYQVVNVADIERLFPEGGDVTIEAL
***************:**:**:**** ***.*: : **:** : :: *
Mflv_5028|M.gilvum_PYR-GCK VAKGLVRKNVLVKVLGDGKLSAKVDITAHKFSGSAREAITAAGGSVTEL-
Mvan_1341|M.vanbaalenii_PYR-1 VAKGLVRKNVLVKVLGDGKLSAKVDVTAHKFSGSAREAITAAGGTATEL-
MSMEG_1474|M.smegmatis_MC2_155 VAKGLVRKNSLVKVLGDGKLTVKVDVTANKFSGSAREAITAAGGSATEL-
TH_0304|M.thermoresistible__bu VAKGAVRKNTLVKVLGDGKLTVKVDVTAHKFSGSAREKITAAGGSATELS
Rv0723|M.tuberculosis_H37Rv VAKGAVRKNALVKVLGDGKLTAKVDVSAHKFSGSARAKITAAGGSATEL-
Mb0744|M.bovis_AF2122/97 VAKGAVRKNALVKVLGDGKLTAKVDVSAHKFSGSARAKITAAGGSATEL-
MAV_4446|M.avium_104 VAKGAVRRNSLVKVLGDGKLTVKVEVSAHKFSGSAREKITAAGGSVTEL-
MMAR_1054|M.marinum_M VAKGAVRKNSLVKVLGDGKLTVKVDVTAHKFSGSAREQITAAGGSVTEL-
MUL_0813|M.ulcerans_Agy99 VAKGAVRKNSLVKVLGDGKLTVKVDITAHKFSGSAREQITAAGGSVTEL-
MLBr_01840|M.leprae_Br4923 VAKKAIRKNSLVKILGDGKLTVKVTLSAHKFSGSARHKITVAGGSVTEL-
MAB_3793c|M.abscessus_ATCC_199 VAKGAVRKNELVKVLGNGDLKVKVSVSANKFSDSAREKITAAGGSINEV-
*** :*:* ***:**:*.*..** ::*:***.*** **.***: .*: