For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDRGDPAHPHGQIYAYPYLTPRTAAMLRQARRHRKRHGDNLFASLLAREVADGSRIVVRGELFTAFVPFA ARWPVEVHIYPNRLVRNLTELNDGELDEFARIYLDVLQRFDRMYSSPLPYMSALHQFSEVQRDGYFHVEL MSIRRSATKLKYLAAAESAMDAFIADVIPESVATRLRELGP
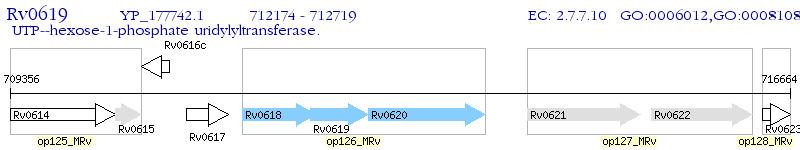
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0619 | galTb | - | 100% (181) | PROBABLE GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE GALTB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0635 | galT | 2e-97 | 99.43% (174) | galactose-1-phosphate uridylyltransferase GALT |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4637 | - | 2e-46 | 49.44% (178) | galactose-1-phosphate uridylyltransferase GalT |
| M. marinum M | MMAR_0940 | galT | 2e-75 | 77.71% (175) | galactose-1-phosphate uridylyltransferase, GalT |
| M. avium 104 | MAV_4565 | galT | 6e-75 | 78.16% (174) | galactose-1-phosphate uridylyltransferase |
| M. smegmatis MC2 155 | MSMEG_3691 | galT | 8e-69 | 72.41% (174) | galactose-1-phosphate uridylyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0691 | galT | 4e-75 | 77.71% (175) | galactose-1-phosphate uridylyltransferase, GalT |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0619|M.tuberculosis_H37Rv --------------------------------------------------
Mb0635|M.bovis_AF2122/97 MSATPPPGGLDASVFIANERGRQLDEALPVGFCVVTAPTRWTLADGRDLL
MAV_4565|M.avium_104 -------------------------------MFDIMGPTRAKLADGRDLL
MMAR_0940|M.marinum_M ----------------------------------MTAPTRAKLADGREVL
MUL_0691|M.ulcerans_Agy99 -------MRRGSHRTPATAGMAAALASVRLVFDIVTAPTRAKLADGREVL
MSMEG_3691|M.smegmatis_MC2_155 ----------------------------------MTQPLRWTLADGREQL
MAB_4637|M.abscessus_ATCC_1997 -----------------------------------MWRTRRELADGREIL
Rv0619|M.tuberculosis_H37Rv --------------------------------------------------
Mb0635|M.bovis_AF2122/97 FFSLPGHVPAPVSDRRPLPERDPA------PSRLRFDRATGQWVIVAAQR
MAV_4565|M.avium_104 FFSLPGHRASPVEDRRPLPPRTAETVAPGGQSQLRFDRSTGQWVIIAALR
MMAR_0940|M.marinum_M FFALPGNVPAPVADRRPLPPRHPH------QSELRFDRSTGQWVIVAALR
MUL_0691|M.ulcerans_Agy99 FFALPGNVPAPVADRRPLPPRHPH------QSELRFDRSTGQWVIVAALR
MSMEG_3691|M.smegmatis_MC2_155 YFSLPGHTPAPVGDRRPLPPRGNQ------QSELRFDRQTGQWVVIAALR
MAB_4637|M.abscessus_ATCC_1997 YFDSIPTERAAVD-RRDLPPVSTQ-------SQIRFDPLLGDWVTLASHR
Rv0619|M.tuberculosis_H37Rv --------------------------------------------------
Mb0635|M.bovis_AF2122/97 QDRTYKPPAARCPLCPGPTGLSSEVPAPDYDVVVFENRFPSLAGAGIAPI
MAV_4565|M.avium_104 QDRTYKPPPDACPLCPGPTGLTSEVPAADYDVVVFENRFPSLSGAAPG-L
MMAR_0940|M.marinum_M QDRTYKPPPDQCPLCPSPTGLTSEVPARDYDVVVFENRFPSLSGSGGP--
MUL_0691|M.ulcerans_Agy99 QDRTYKPPPDQCPLCPSPTGLTSEVPARDYDVVVFENRFPSLSGSGGP--
MSMEG_3691|M.smegmatis_MC2_155 QDRTYLPAADQCPLCPGPTGETSEVPAPDYDVVVFENRFPSLSGGGEFTL
MAB_4637|M.abscessus_ATCC_1997 QTRTFLPPTDLCPLCPSTPERPTEIPEHAYEVVAFENRFPSLS-------
Rv0619|M.tuberculosis_H37Rv --------------------------------------------------
Mb0635|M.bovis_AF2122/97 GAPDGDGFVSAPGHGRCEVICFSADHTGSFAGLDPAHAGLVVHAWRHRTA
MAV_4565|M.avium_104 PAAGADGFVSAPAHGRCEVVCFSADHTGSFAGLPPAHARLVVQAWRHRTA
MMAR_0940|M.marinum_M -PPGGDGFVSAPGDGRCEVICFSSDHGGSFADLPAPHARLVIEAWRHRTG
MUL_0691|M.ulcerans_Agy99 -PPGGDGFVSAPGDGRCEVICFSSDHGGSFADLPAPHARLVIEAWRHRTG
MSMEG_3691|M.smegmatis_MC2_155 PDTAGAEFLSAPGHGRCEVICFSSDHTGAFSQLPLPHARLVVSAWRHRTA
MAB_4637|M.abscessus_ATCC_1997 LHAQAATPPAGAGFGRCEVLCFTSEHDTSFAQLEPRRARLIVDVWAERTK
Rv0619|M.tuberculosis_H37Rv --------------GDRGD-----PAHPHGQIYAYPYLTPRTAAMLRQAR
Mb0635|M.bovis_AF2122/97 ELTALPGVAQVFCFENRGEEIGVTLTHPHGQIYAYPYLTPRTAAMLRQAR
MAV_4565|M.avium_104 ELMATPGIEQVFCFENRGEEIGVTLTHPHGQIYGYPYLTPRTAAMLGQAD
MMAR_0940|M.marinum_M DLIGQPGIEQVFCFENRGEEIGVTLTHPHGQIYGYPFLTPRTSVMLEQAG
MUL_0691|M.ulcerans_Agy99 DLIGQPGIEQVFCFENRGEEIGVTLTHPHGQIYGYPLLTPRTSVMLEQAG
MSMEG_3691|M.smegmatis_MC2_155 DLLSRNGIEQVFCFENRGEAIGVTLNHPHGQIYGYPYLTPRTEQMLRVAG
MAB_4637|M.abscessus_ATCC_1997 ELSAIDGIEQVHCFENHGEEIGVTLAHPHGQIYAYPFITAKTARVLSNVD
::*: *******.** :*.:* :* .
Rv0619|M.tuberculosis_H37Rv RHRKRHGDNLFASLLAREVADGSRIVVRGELFTAFVPFAARWPVEVHIYP
Mb0635|M.bovis_AF2122/97 RHRKRHGDNLFASLLAREVADGSRIVVRGELFTAFVPFAARWPVEVHIYP
MAV_4565|M.avium_104 DHRKRHGGNLFGDLLAREVADGSRIIARDDLFTAFVPFAARWPVEVHIYP
MMAR_0940|M.marinum_M RHRMRHGCNLFEDLLAREVADGSRIVARGDRFTAFVPFAARWPVEVHIYP
MUL_0691|M.ulcerans_Agy99 RHRMRHGCNLFEDLLAREVADGSRIVARGDRFTAFVPFAARWPVEVHIYP
MSMEG_3691|M.smegmatis_MC2_155 EHRARNGTNLFADLLARERADSVRVVAQTDSFTAFVPFAARWPVEVHIYP
MAB_4637|M.abscessus_ATCC_1997 THRKQHGTNLFGEILAAERATGTRVVTANDEWTAFVPSFARWPYEVSLYP
** ::* *** .:** * * . *::. : :***** **** ** :**
Rv0619|M.tuberculosis_H37Rv NRLVRNLTELNDGELDEFARIYLDVLQRFDRMYSSPLPYMSALHQFS---
Mb0635|M.bovis_AF2122/97 NRLVRNLTELNDGELDEFARIYLDVLQRFDRMYSSPLPYMSALHQFS---
MAV_4565|M.avium_104 NRFVPNLTALRDDEVDGFVAVYLDVLRRFDRMYDAPLPYISALHQYTGT-
MMAR_0940|M.marinum_M NRFVRNLVDLDAAECNEFTEIYLDVLRRFDRMYSGPLPYISALHQYRDT-
MUL_0691|M.ulcerans_Agy99 NRFVRNLADLDAAECNEFTEIYLDVLRRFDRMYSGPLPYISALHQYRDT-
MSMEG_3691|M.smegmatis_MC2_155 NRFAHSLLDLDDSELDEFTEIYRDVLQRFDRMYPTELPYISALHQYRDS-
MAB_4637|M.abscessus_ATCC_1997 HRQVADIPDLDDAARSAFAELYLDVLQRFARRFDNPMPYIASWNQAPTVA
:* . .: * . *. :* ***:** * : :**::: :*
Rv0619|M.tuberculosis_H37Rv --EVQRDGYFHVELMSIRRSATKLKYLAAAESAMDAFIADVIPESVATRL
Mb0635|M.bovis_AF2122/97 --EVQRDGYFHVELMSIRRSATKLKYLAAAESAMDAFIADVIPESVAARL
MAV_4565|M.avium_104 --GAQAHGYFHVELMSIRRSATKLKFLAASESAMDAFISDVTPESVAQRL
MMAR_0940|M.marinum_M --EAQREGYFHVELMSIRRSATALKYLAASESAMGAFISDVTPETVAERL
MUL_0691|M.ulcerans_Agy99 --EAQREGYFHVELMSIRRSATALKYLAASESAMGAFISDVTPETVAERL
MSMEG_3691|M.smegmatis_MC2_155 --ERQREGYFHVELMSVRRSENKLKYLAGSESAMDAFISDVTPESVAQRL
MAB_4637|M.abscessus_ATCC_1997 SGAIRDNWWLHLQVFSIQRAPRKLKYLAGSESGMGAFISDTTPEQVAAEL
: . ::*::::*::*: **:**.:**.*.***:*. ** ** .*
Rv0619|M.tuberculosis_H37Rv RELGP
Mb0635|M.bovis_AF2122/97 RELGP
MAV_4565|M.avium_104 RELR-
MMAR_0940|M.marinum_M RELG-
MUL_0691|M.ulcerans_Agy99 RELG-
MSMEG_3691|M.smegmatis_MC2_155 REV--
MAB_4637|M.abscessus_ATCC_1997 RDVI-
*::